| Glycine, serine and threonine metabolism | 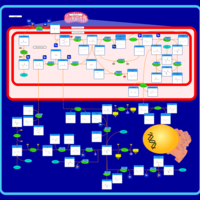 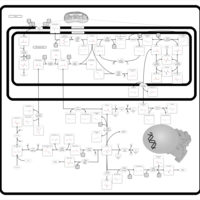 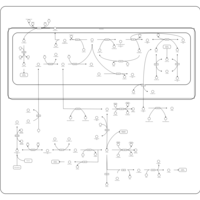 |
| Transcription/Translation |    |
| Dihydropyrimidine Dehydrogenase Deficiency (DHPD) | 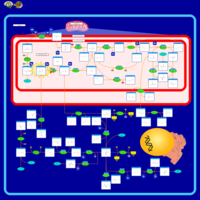 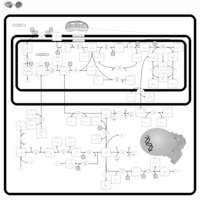 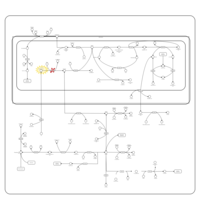 |
| Non Ketotic Hyperglycinemia | 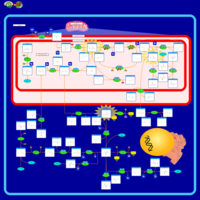 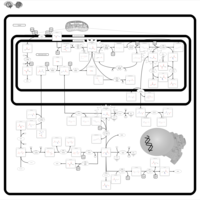 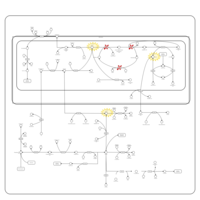 |
| Dimethylglycine Dehydrogenase Deficiency | 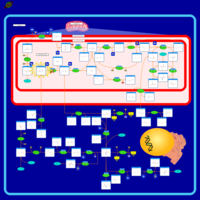 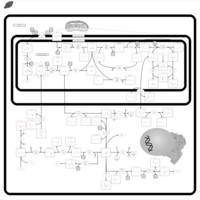  |
| Sarcosinemia | 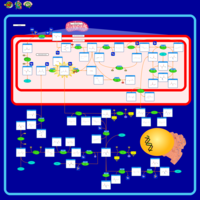 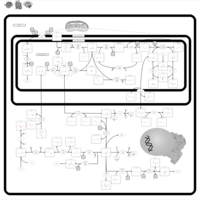 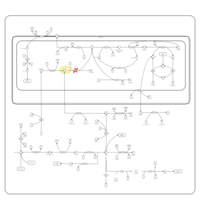 |
| Azithromycin Action Pathway | 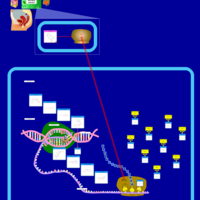 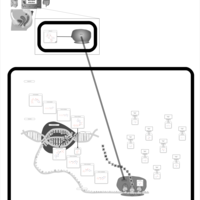 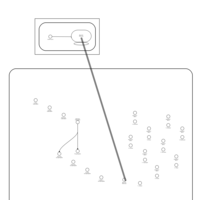 |
| Clarithromycin Action Pathway | 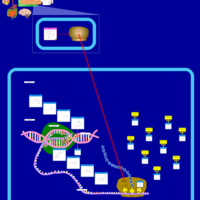 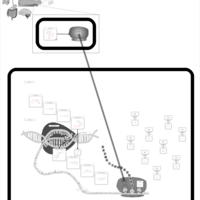 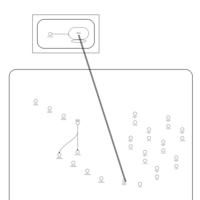 |
| Clindamycin Action Pathway | 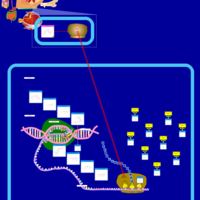 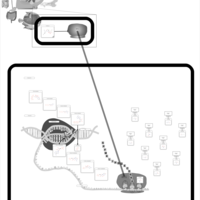  |
| Erythromycin Action Pathway | 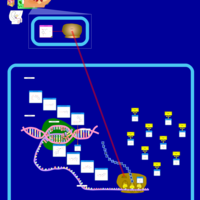 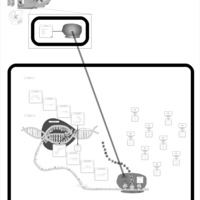 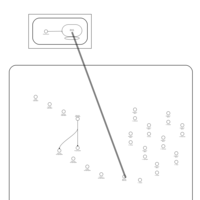 |
| Roxithromycin Action Pathway | 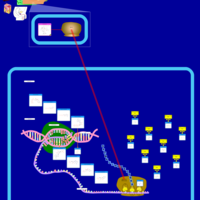 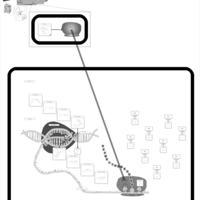  |
| Telithromycin Action Pathway | 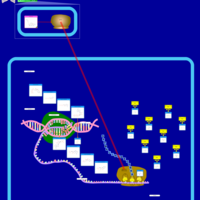   |
| Amikacin Action Pathway | 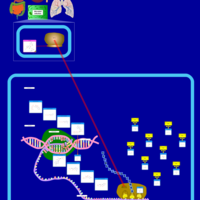 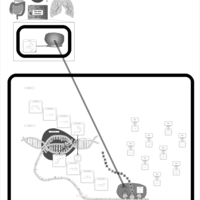  |
| Gentamicin Action Pathway | 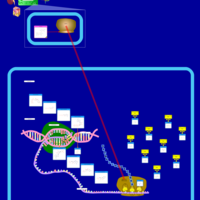 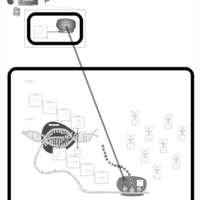 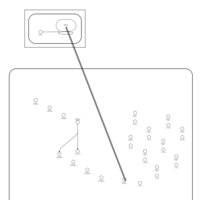 |
| Kanamycin Action Pathway | 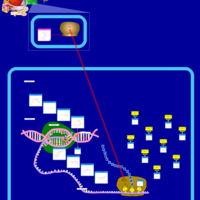 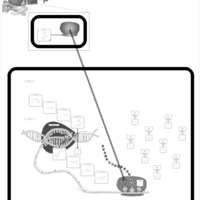  |
| Neomycin Action Pathway |    |
| Netilmicin Action Pathway |   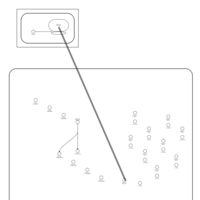 |
| Spectinomycin Action Pathway | 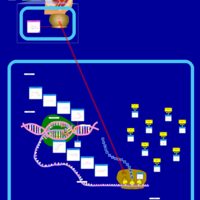 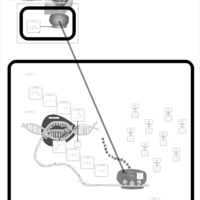 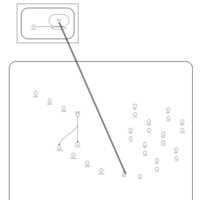 |
| Streptomycin Action Pathway | 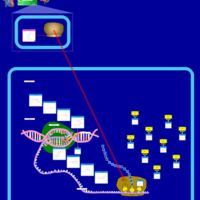   |
| Clomocycline Action Pathway | 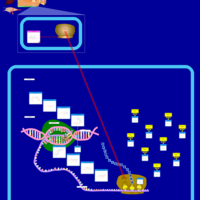 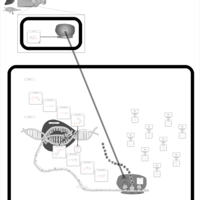 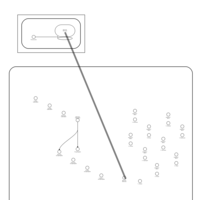 |
| Demeclocycline Action Pathway | 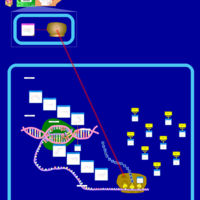 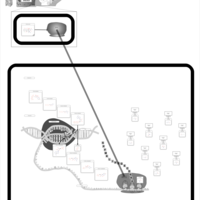  |
| Doxycycline Action Pathway | 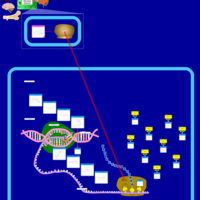 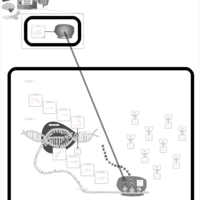  |
| Minocycline Action Pathway | 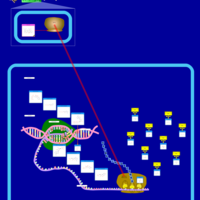   |
| Oxytetracycline Action Pathway | 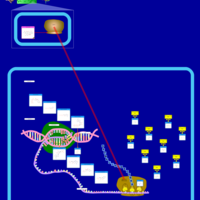   |
| Tetracycline Action Pathway | 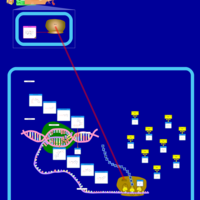   |
| Lymecycline Action Pathway | 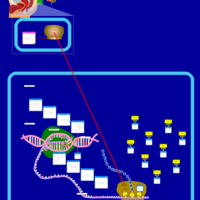 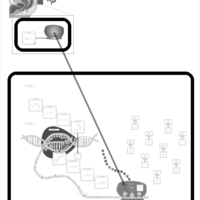 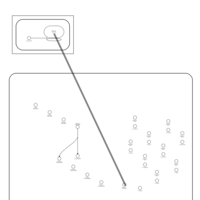 |
| Dimethylglycine Dehydrogenase Deficiency |    |
| Hyperglycinemia, non-ketotic |    |
| Tobramycin Action Pathway | 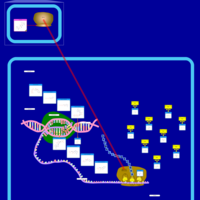 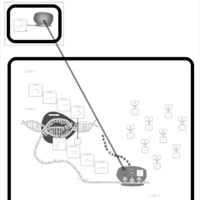  |
| Tigecycline Action Pathway |    |
| Arbekacin Action Pathway | 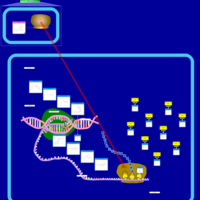 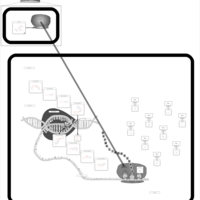  |
| Paromomycin Action Pathway | 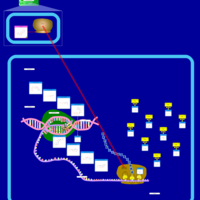   |
| 3-Phosphoglycerate dehydrogenase deficiency | 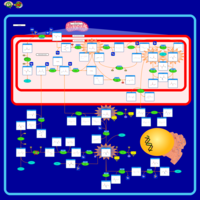 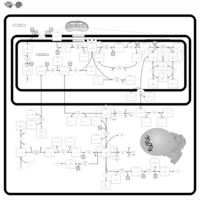 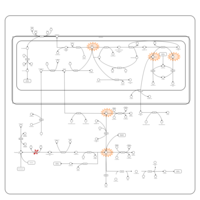 |
| Rolitetracycline Action Pathway | 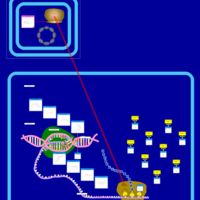 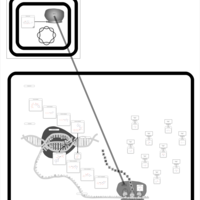  |
| Methacycline Action Pathway | 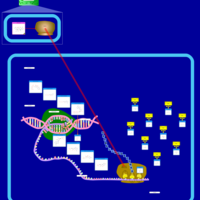   |
| Lincomycin Action Pathway | 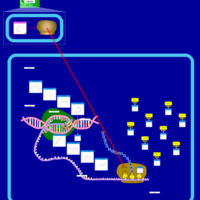 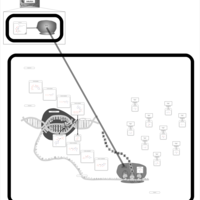 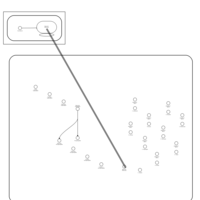 |
| Chloramphenicol Action Pathway | 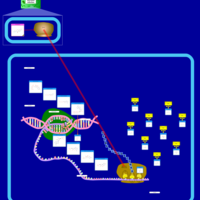 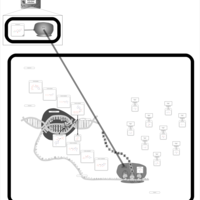  |
| Troleandomycin Action Pathway |    |
| Josamycin Action Pathway | 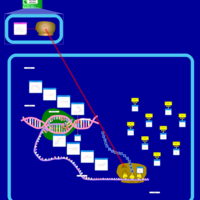   |