Instructions
Annotate
Step 1:
Upload a full bacterial genome for BASys2 to annotate. There are two options:
Click the green "Load Example" button under your section of choice if you do not have input. Or you can download an example Genbank file or example Fasta file by clicking on the links.
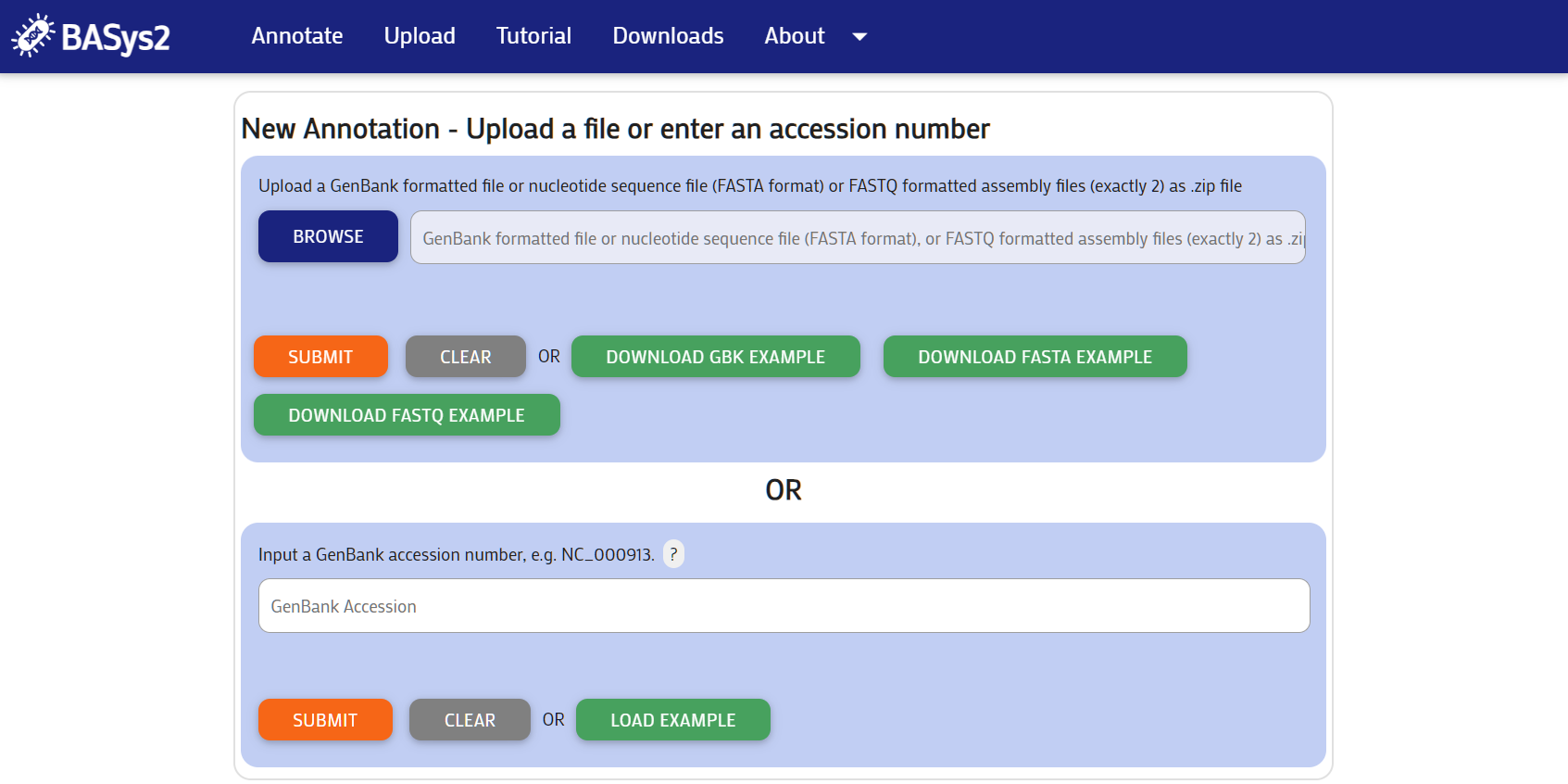
Step 2:
Press the "Submit" button. The following page will be shown. Bookmark the page or save the URL to access your results later.
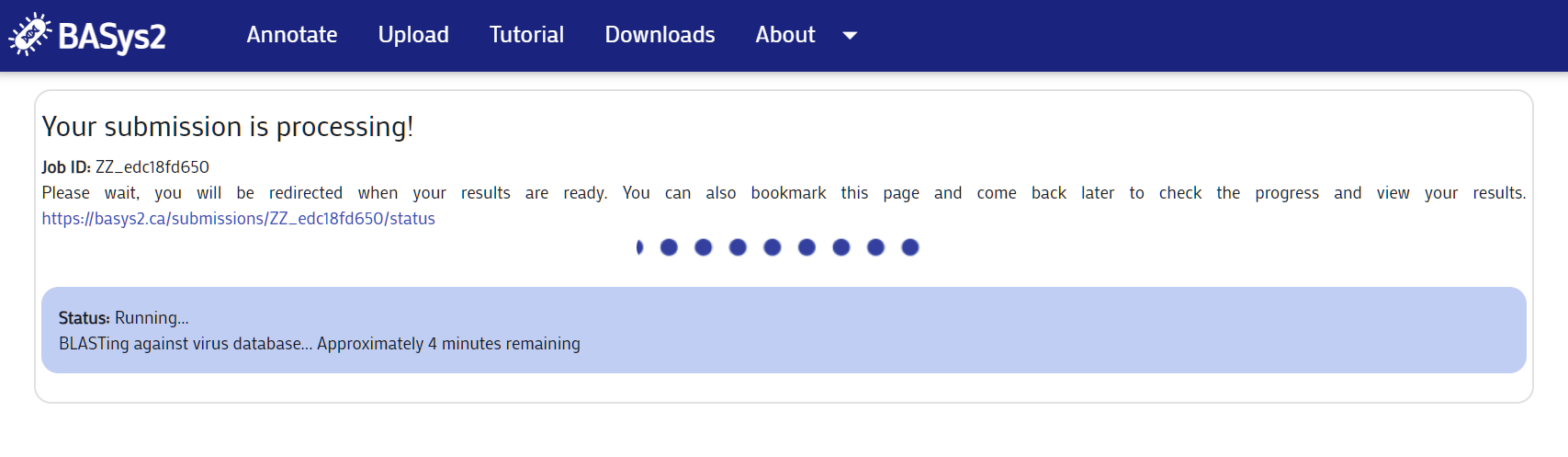
Step 3:
Once the file upload has been completed, the following results page will be shown. You may download the genome map by clicking the "BASYS2-MAP.json" download link. This file can be uploaded to the BASys2 Visualizer Software that can be downloaded HERE. You may also download the images of the genome map by clicking on thefile_downloadicon below the viewers. The entire results package may be downloaded via the 'Download Results' link at the top of the results section. You may view detailed OUTPUT INFORMATION here.
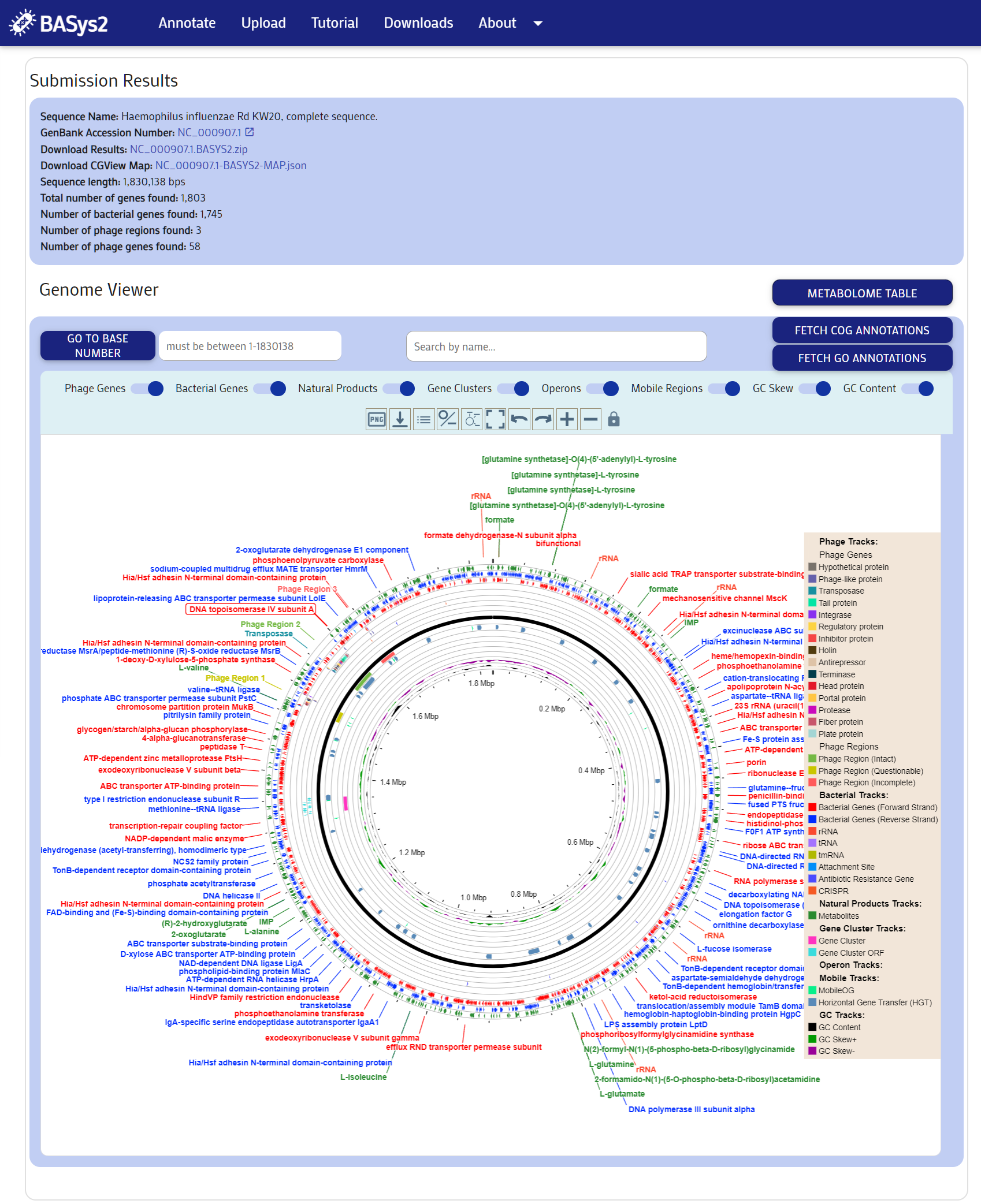
Step 4:
You can explore the genome map using the map controls and clicking on genes. Clicking on a gene will display a pop-up with the gene's details. You may download a FASTA file of the selected gene by clicking the "Download FASTA" button.
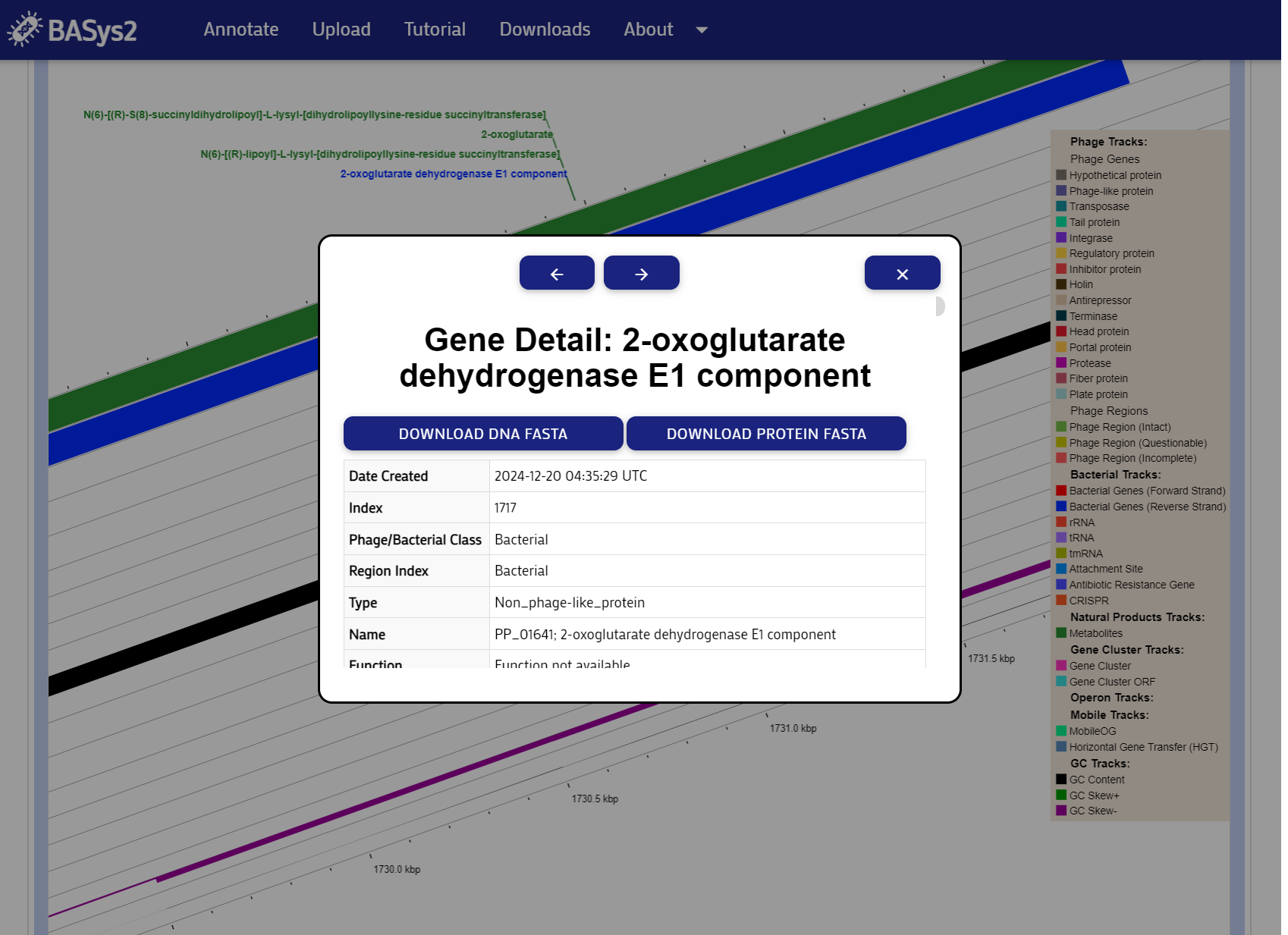
Step 5:
Clicking the "Metabolome Table" button above the genome map will take you to the Metabolome page. Here you can view the full metabolome of the organism. You can filter the list by entering a search term in the search bar. You can also sort the list by clicking on the column headers.
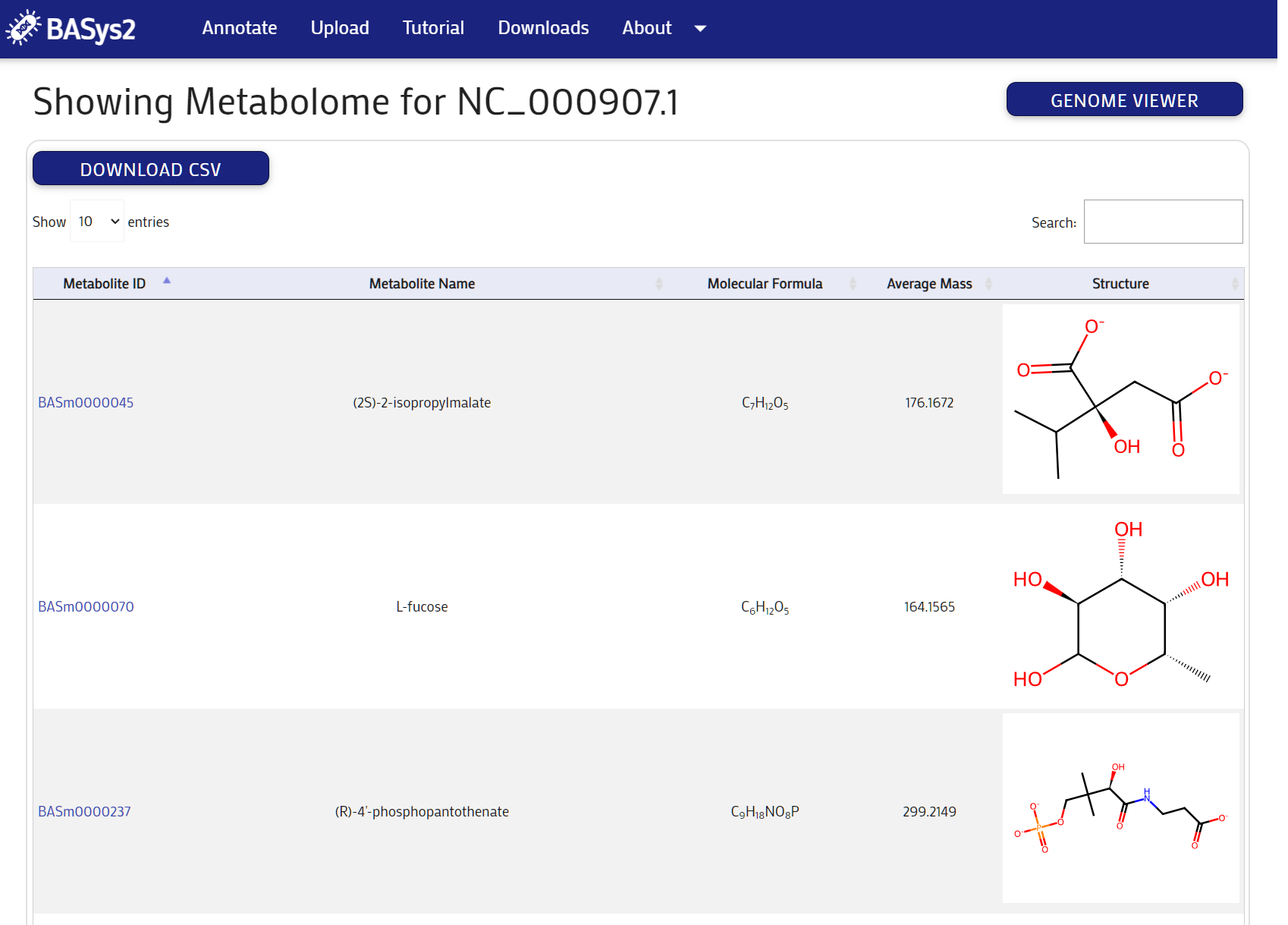
Step 6:
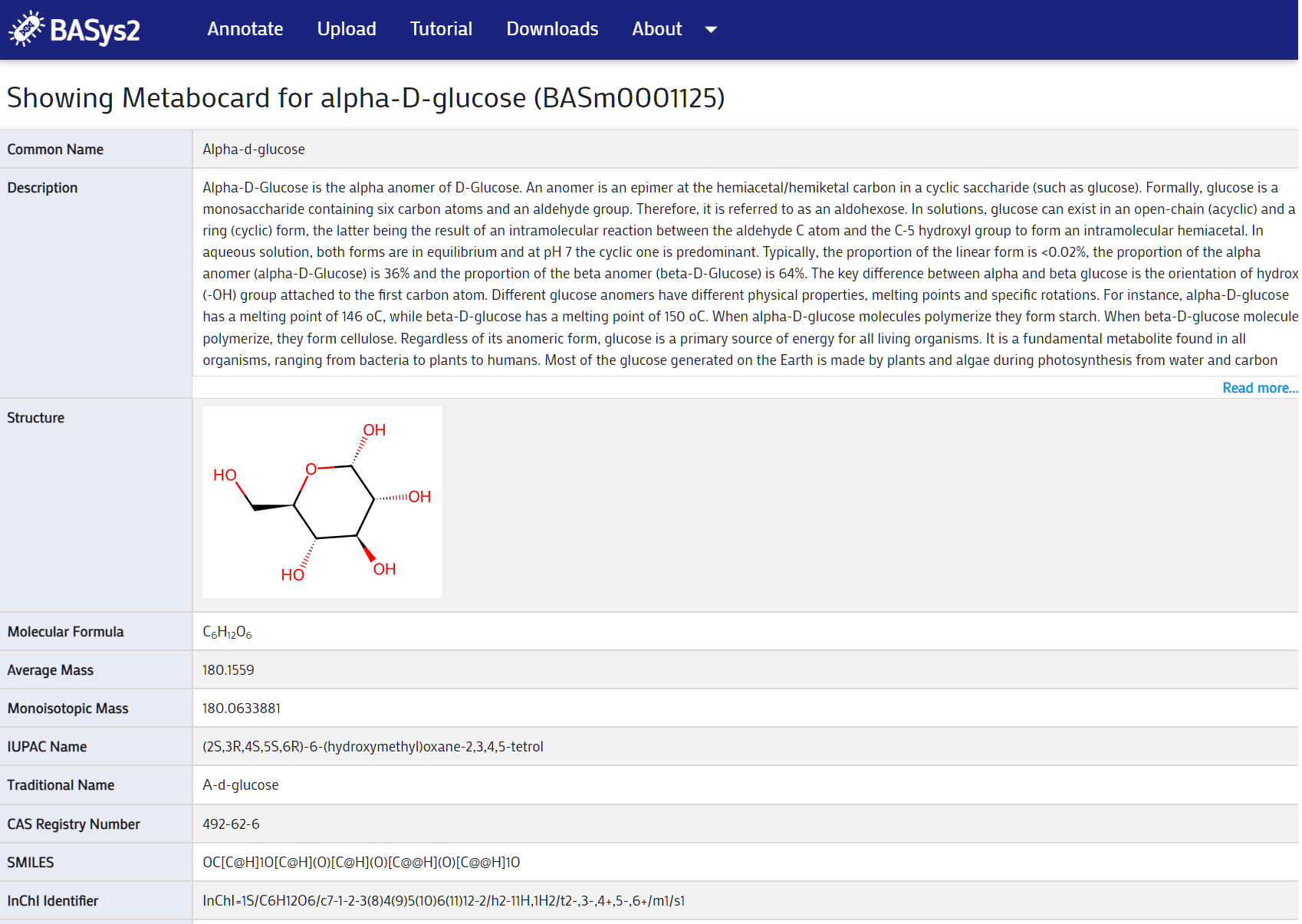
Clicking on a the metabolite ID link will take you to the Metabolite page. Here you can view the details of the metabolite.
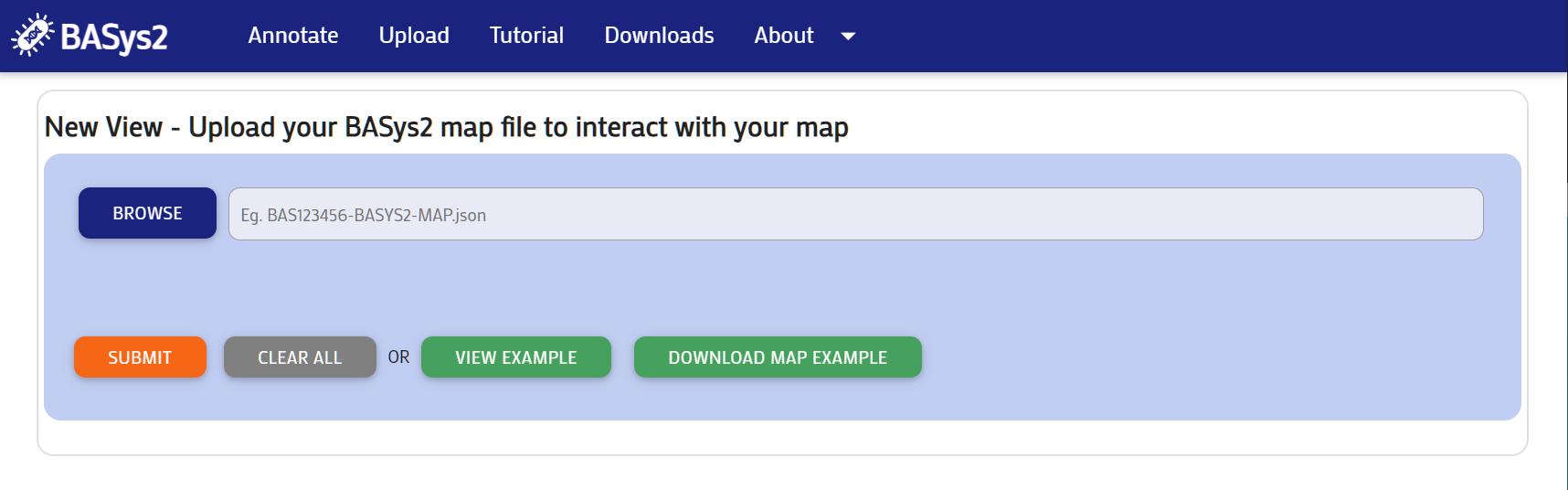
Upload
Step 1:
Upload a map file from a previously run BASys2 job. This is the same process as uploading into the downloadable BASys2 Visualizer.
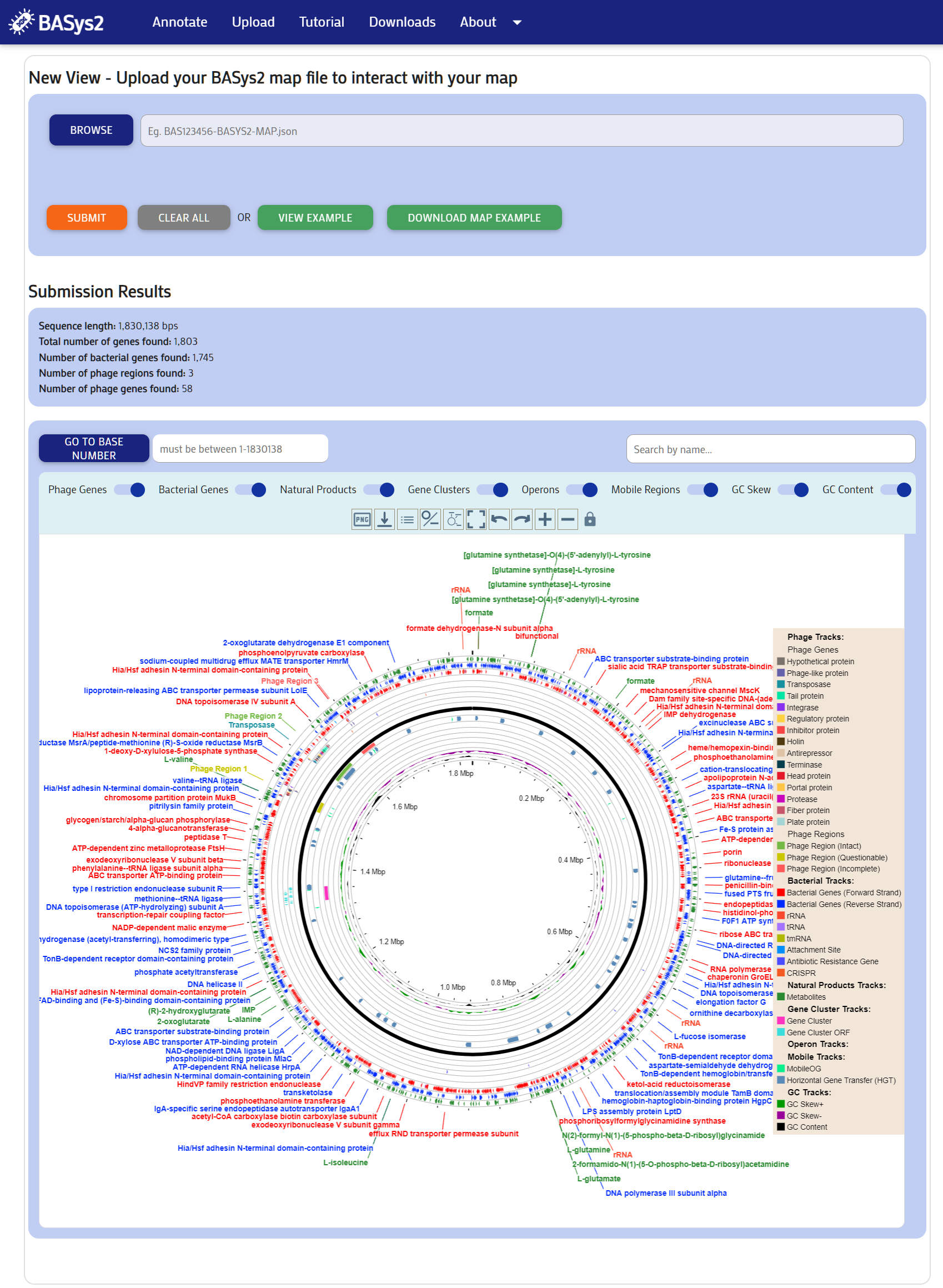
Step 2:
You can explore the genome map in the same way as in the Annotate section.
Browser & Operating System Compatibility
BASys2 has been tested on all major browsers (Firefox, Google Chrome, Microsoft Edge and Safari) on MacOS, Linux, and Windows.
BASys2 Visualizer has been tested on all on MacOS, Linux, and Windows.
Disclaimer
BASys2 is a predictive tool, and positions of the predicted prophage regions and/or attachment sites may differ from actual prophage regions and/or attachment sites in some cases.