| Purine metabolism |  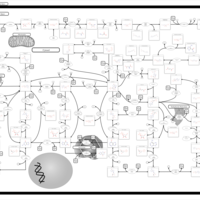 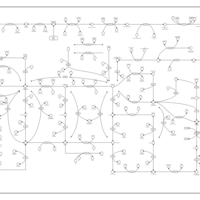 |
| Fructose and mannose metabolism | 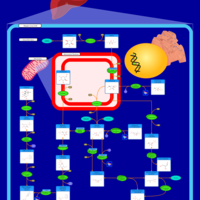 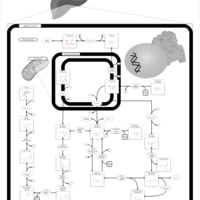 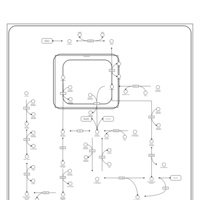 |
| Pyruvate metabolism |  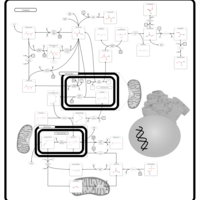  |
| Pterine Biosynthesis | 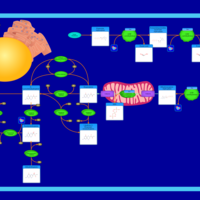 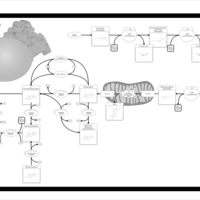 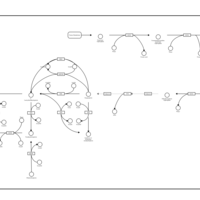 |
| Transcription/Translation |    |
| Citric Acid Cycle | 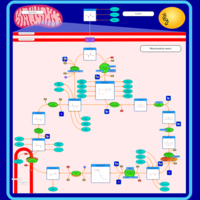 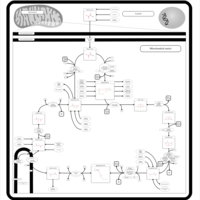 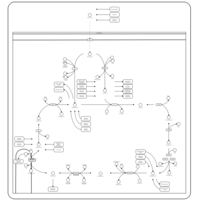 |
| Aspartate Metabolism | 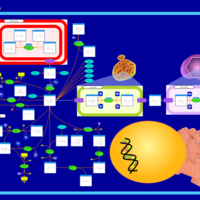 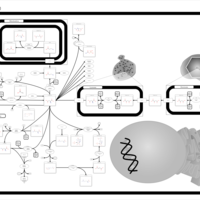 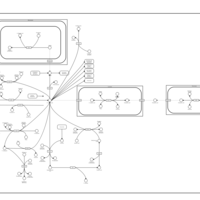 |
| Gluconeogenesis | 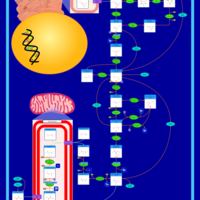 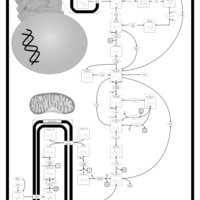 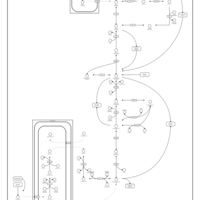 |
| Adenosine Deaminase Deficiency |  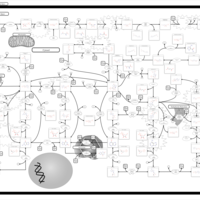 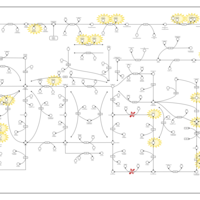 |
| Adenylosuccinate Lyase Deficiency | 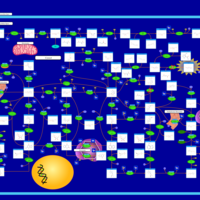 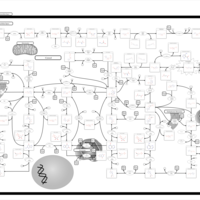 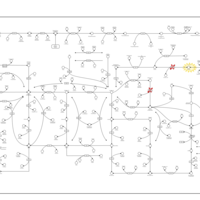 |
| AICA-Ribosiduria | 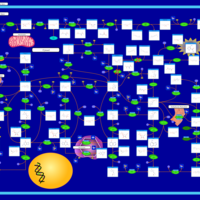 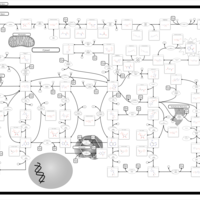 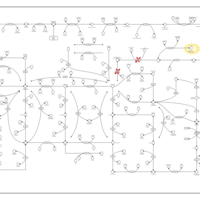 |
| Canavan Disease | 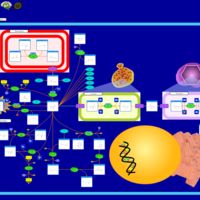 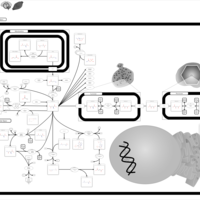 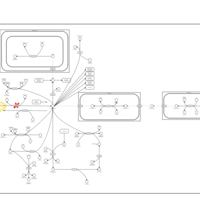 |
| Hypoacetylaspartia | 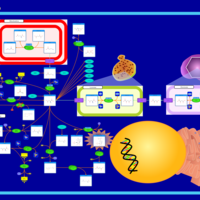 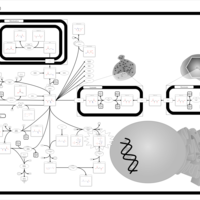  |
| Leigh Syndrome | 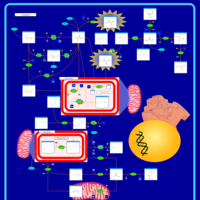 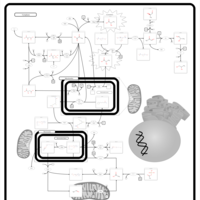 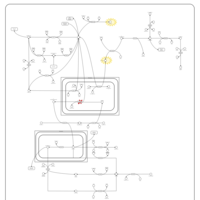 |
| Molybdenum Cofactor Deficiency |    |
| Purine Nucleoside Phosphorylase Deficiency |    |
| Pyruvate Dehydrogenase Complex Deficiency | 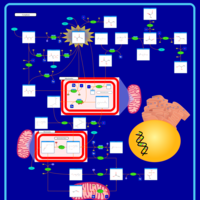 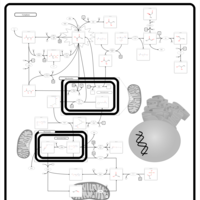 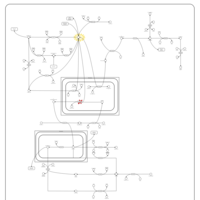 |
| Xanthine Dehydrogenase Deficiency (Xanthinuria) | 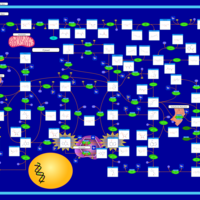 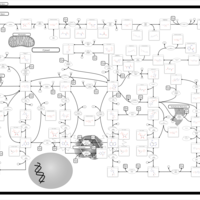  |
| Azithromycin Action Pathway | 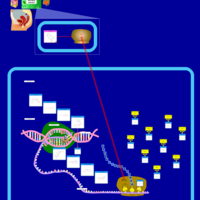 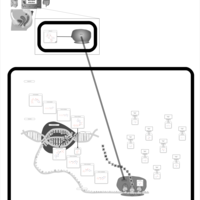 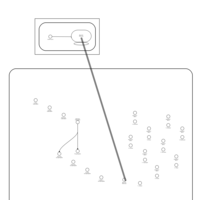 |
| Clarithromycin Action Pathway | 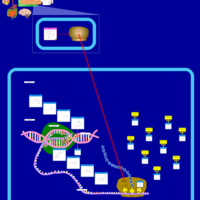 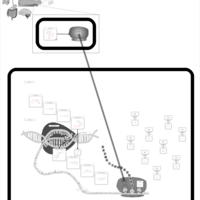  |
| Clindamycin Action Pathway |    |
| Erythromycin Action Pathway | 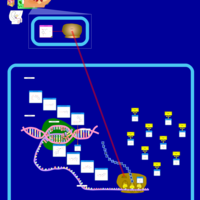   |
| Roxithromycin Action Pathway | 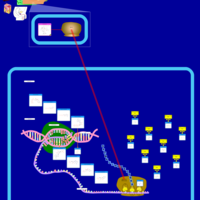   |
| Telithromycin Action Pathway | 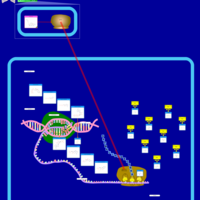  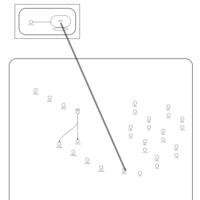 |
| Amikacin Action Pathway | 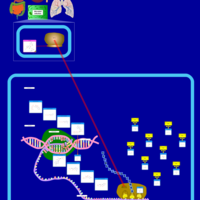 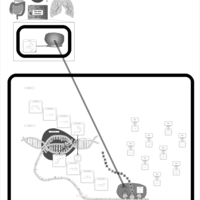 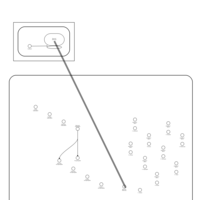 |
| Gentamicin Action Pathway | 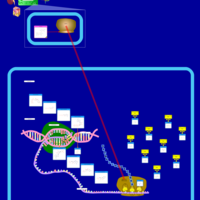 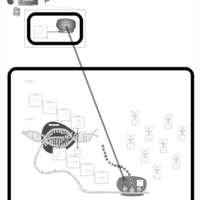 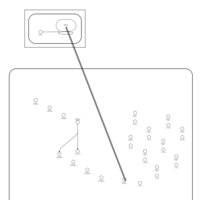 |
| Kanamycin Action Pathway | 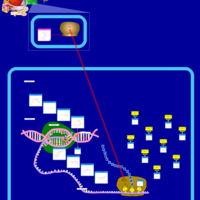 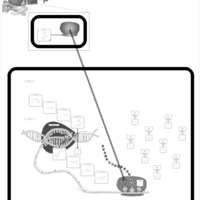 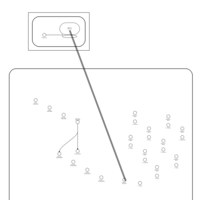 |
| Neomycin Action Pathway | 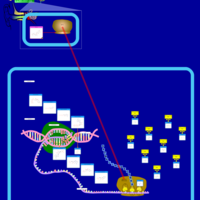  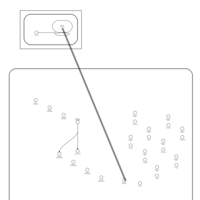 |
| Netilmicin Action Pathway |    |
| Spectinomycin Action Pathway |    |
| Streptomycin Action Pathway |    |
| Clomocycline Action Pathway | 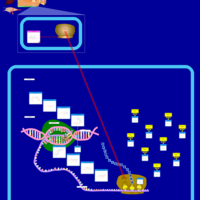   |
| Demeclocycline Action Pathway | 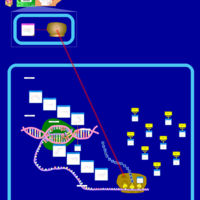   |
| Doxycycline Action Pathway |   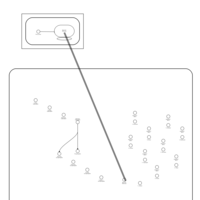 |
| Minocycline Action Pathway | 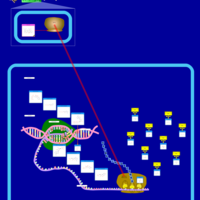 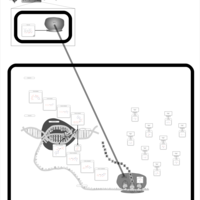  |
| Oxytetracycline Action Pathway | 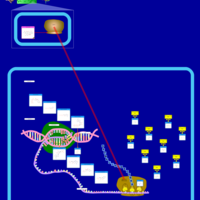  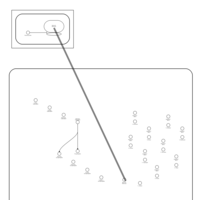 |
| Tetracycline Action Pathway | 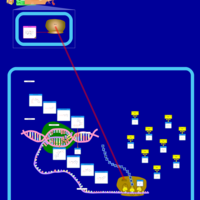 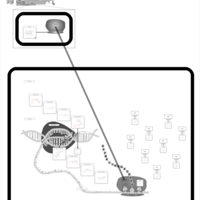 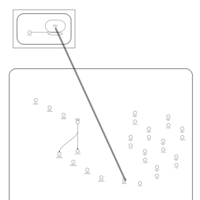 |
| Lymecycline Action Pathway |    |
| Dopamine Activation of Neurological Reward System |    |
| Excitatory Neural Signalling Through 5-HTR 4 and Serotonin | 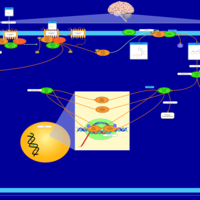 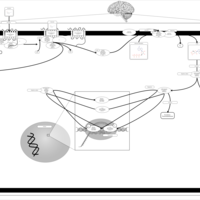 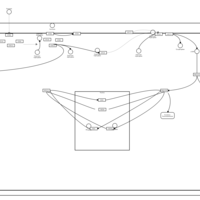 |
| Corticotropin Activation of Cortisol Production | 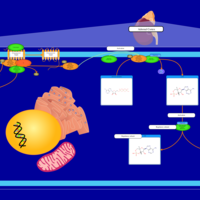 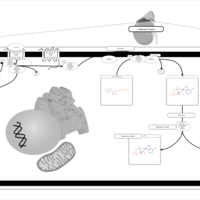 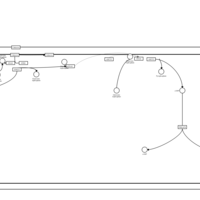 |
| Excitatory Neural Signalling Through 5-HTR 7 and Serotonin | 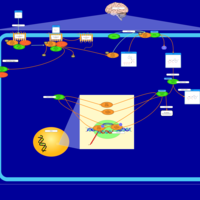 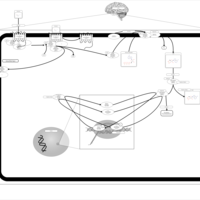 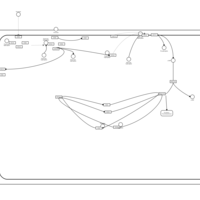 |
| Excitatory Neural Signalling Through 5-HTR 6 and Serotonin | 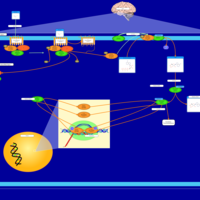 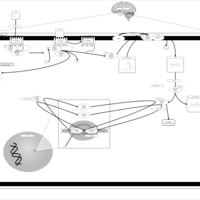 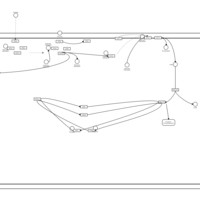 |
| Intracellular Signalling Through Adenosine Receptor A2a and Adenosine |  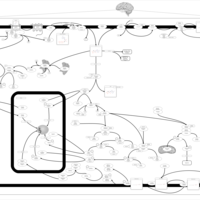 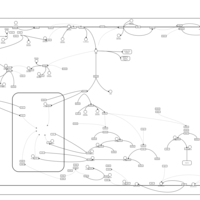 |
| Intracellular Signalling Through Adenosine Receptor A2b and Adenosine | 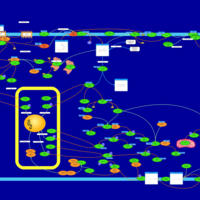 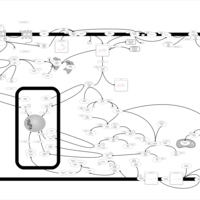 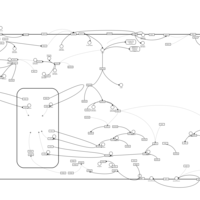 |
| Vasopressin Regulation of Water Homeostasis | 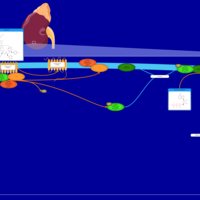 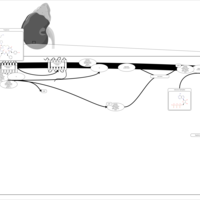 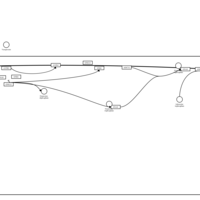 |
| Intracellular Signalling Through FSH Receptor and Follicle Stimulating Hormone | 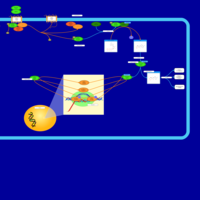 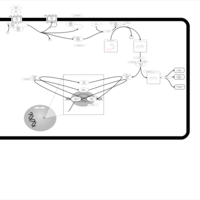 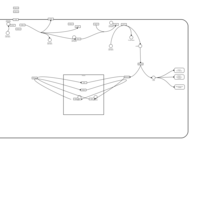 |
| Pyruvate Decarboxylase E1 Component Deficiency (PDHE1 Deficiency) | 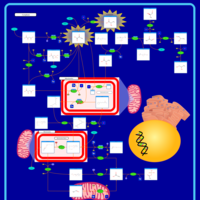 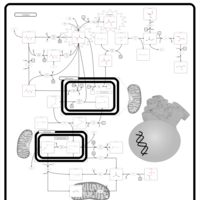 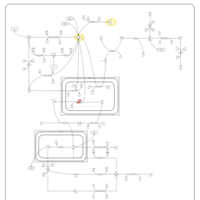 |
| Intracellular Signalling Through Histamine H2 Receptor and Histamine | 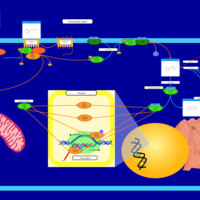 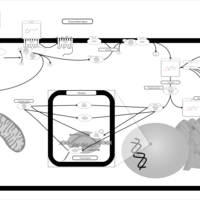  |
| Intracellular Signalling Through LHCGR Receptor and Luteinizing Hormone/Choriogonadotropin |    |
| Intracellular Signalling Through PGD2 receptor and Prostaglandin D2 | 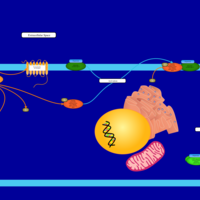 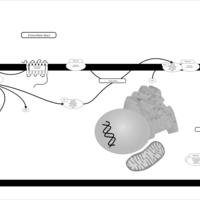  |
| Intracellular Signalling Through Prostacyclin Receptor and Prostacyclin | 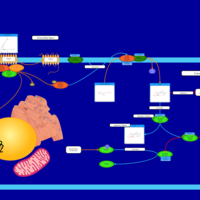 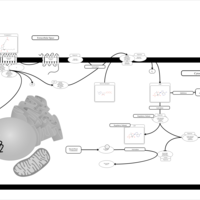 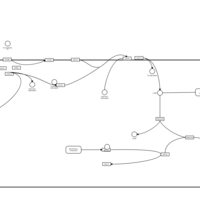 |
| Lesch-Nyhan Syndrome (LNS) | 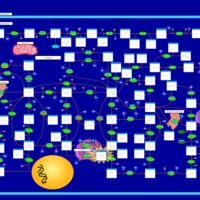 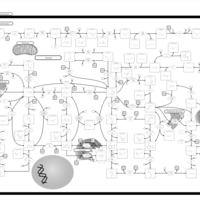 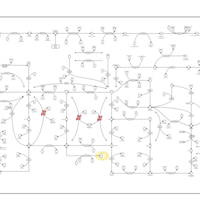 |
| Gout or Kelley-Seegmiller Syndrome | 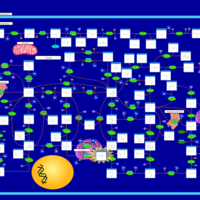 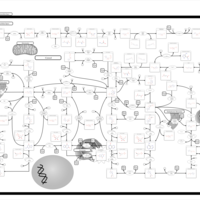 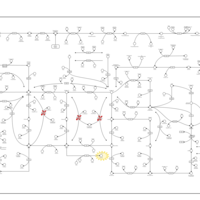 |
| Glycogen Storage Disease Type 1A (GSD1A) or Von Gierke Disease | 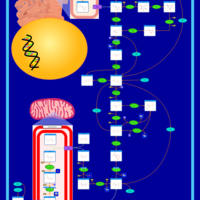 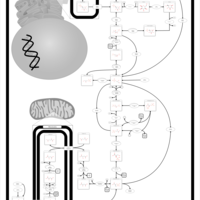 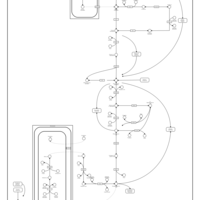 |
| Insulin Signalling | 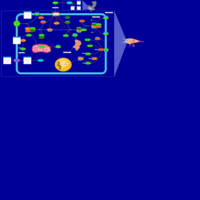 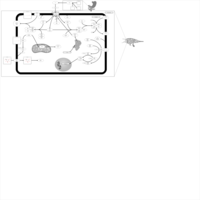 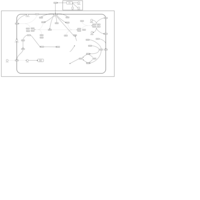 |
| Azathioprine Action Pathway | 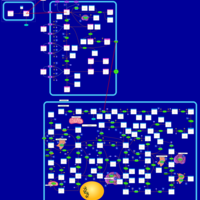 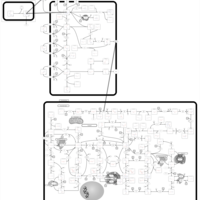 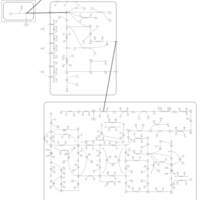 |
| Mercaptopurine Action Pathway |  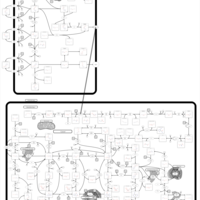 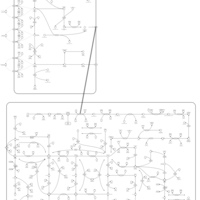 |
| Thioguanine Action Pathway | 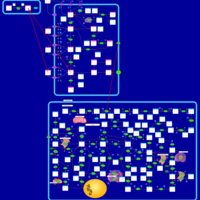  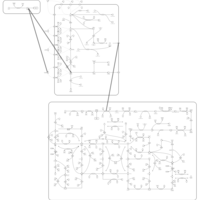 |
| DNA Replication Fork |    |
| Dopa-responsive dystonia | 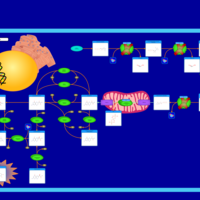 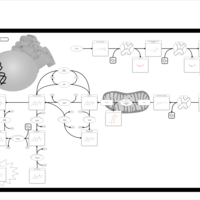 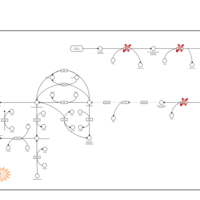 |
| Hyperphenylalaniemia due to guanosine triphosphate cyclohydrolase deficiency | 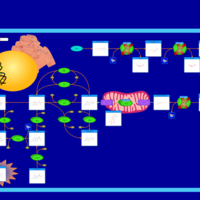   |
| Hyperphenylalaninemia due to 6-pyruvoyltetrahydropterin synthase deficiency (ptps) | 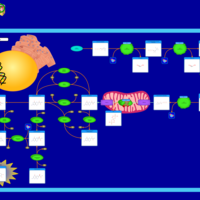 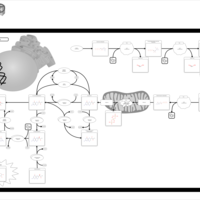 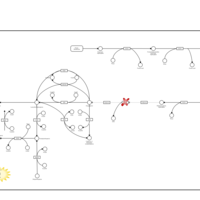 |
| Hyperphenylalaninemia due to dhpr-deficiency | 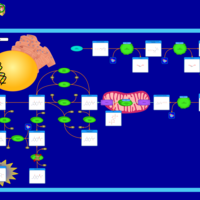 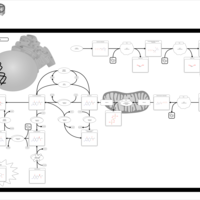 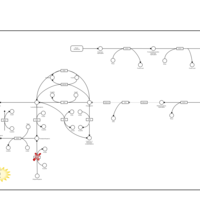 |
| Segawa syndrome | 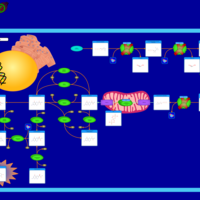 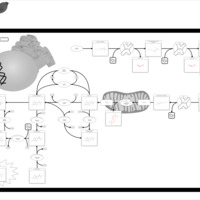 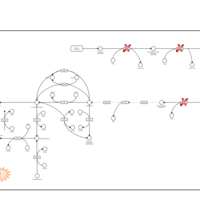 |
| Sepiapterin reductase deficiency | 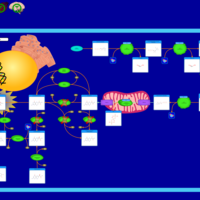 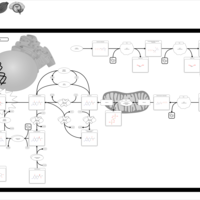 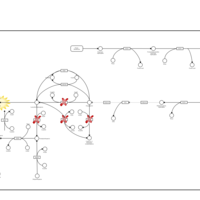 |
| Xanthinuria type I |  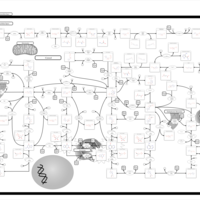 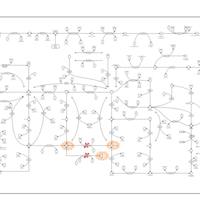 |
| Xanthinuria type II | 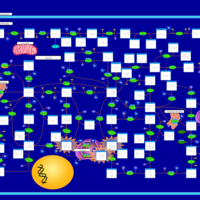 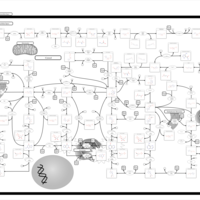  |
| Adenine phosphoribosyltransferase deficiency (APRT) | 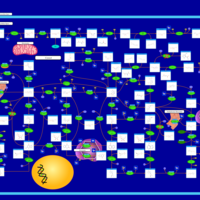 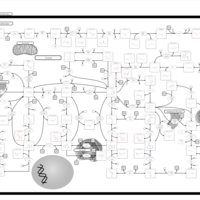 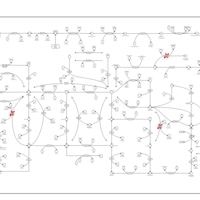 |
| Mitochondrial DNA depletion syndrome |    |
| Myoadenylate deaminase deficiency |    |
| Congenital lactic acidosis | 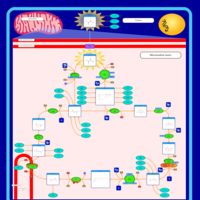 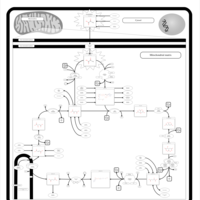 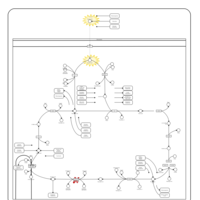 |
| Fumarase deficiency |  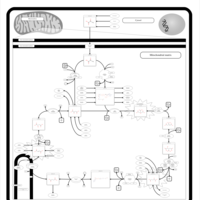 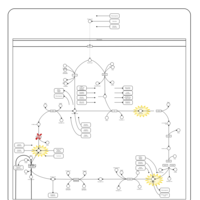 |
| Mitochondrial complex II deficiency | 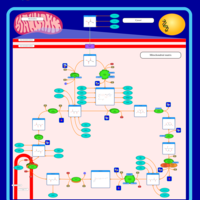 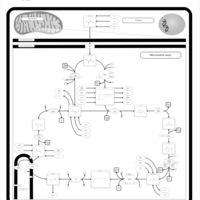 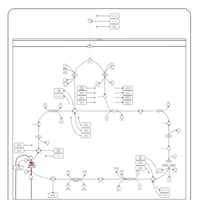 |
| 2-ketoglutarate dehydrogenase complex deficiency | 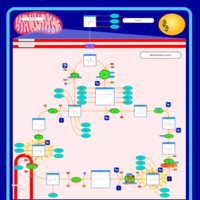 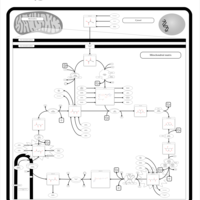 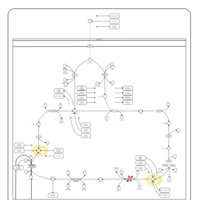 |
| Pyruvate dehydrogenase deficiency (E3) |  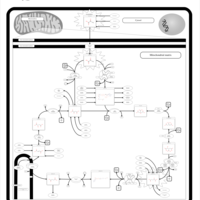 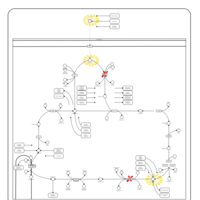 |
| Pyruvate dehydrogenase deficiency (E2) | 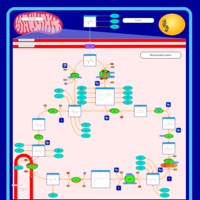 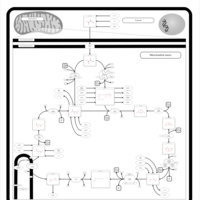 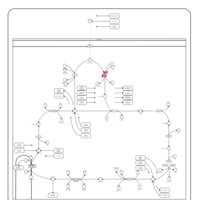 |
| Primary hyperoxaluria II, PH2 | 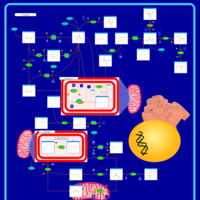 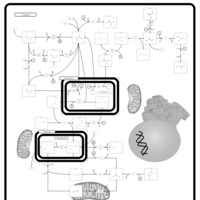 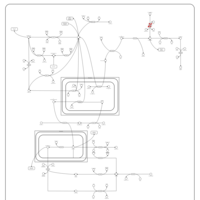 |
| Pyruvate kinase deficiency | 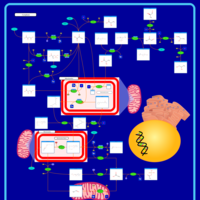 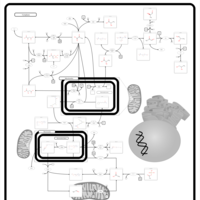 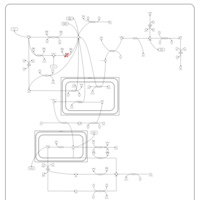 |
| Phosphoenolpyruvate carboxykinase deficiency 1 (PEPCK1) | 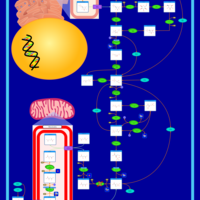 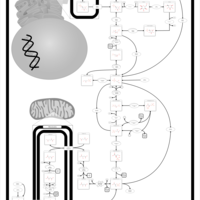 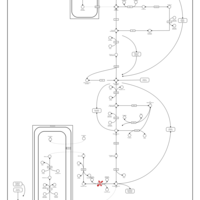 |
| Fructosuria | 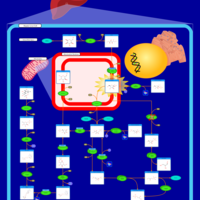 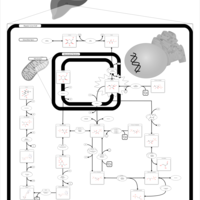 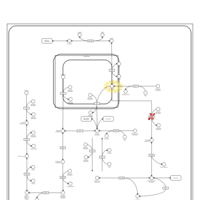 |
| Fructose-1,6-diphosphatase deficiency | 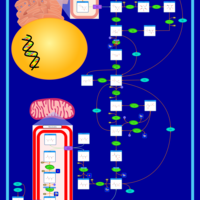 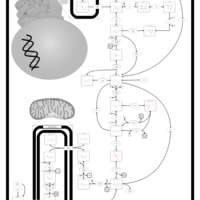 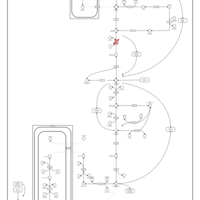 |
| Triosephosphate isomerase | 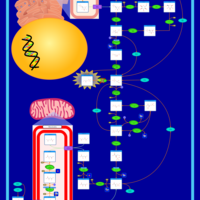 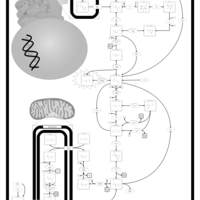 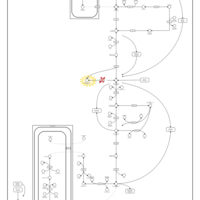 |
| Glycogenosis, Type IB |    |
| Glycogenosis, Type IC |    |
| Glycogenosis, Type IA. Von gierke disease |    |
| Warburg Effect | 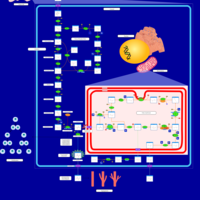 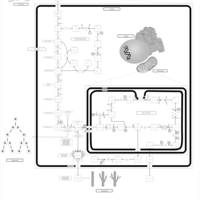 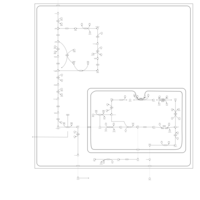 |
| Tobramycin Action Pathway | 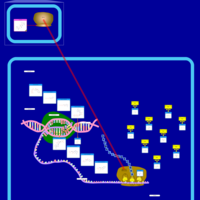 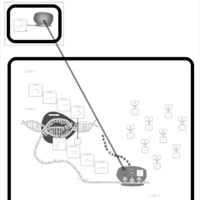 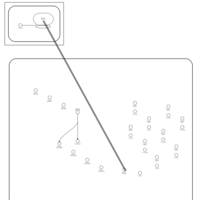 |
| Tigecycline Action Pathway | 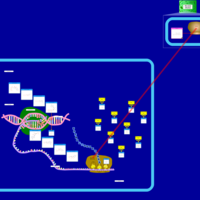   |
| Arbekacin Action Pathway | 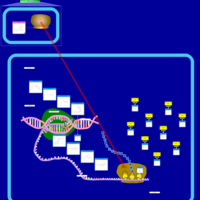   |
| Paromomycin Action Pathway |    |
| Fructose intolerance, hereditary | 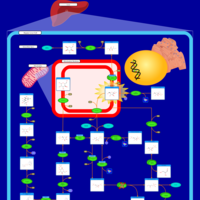 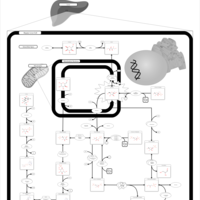 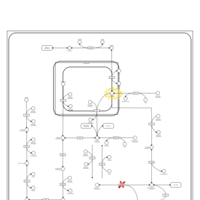 |
| Rolitetracycline Action Pathway | 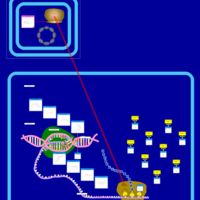   |
| Methacycline Action Pathway | 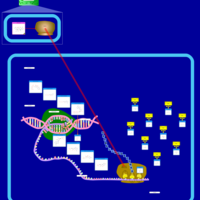   |
| Lincomycin Action Pathway |    |
| Chloramphenicol Action Pathway |    |
| Troleandomycin Action Pathway | 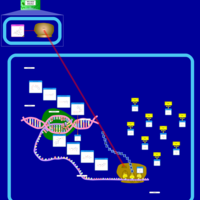   |
| Josamycin Action Pathway | 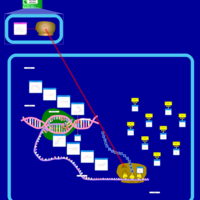   |
| Activation of PKC through G protein coupled receptor |  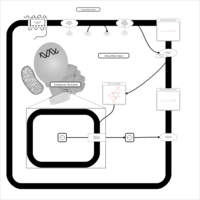 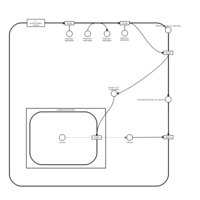 |
| The oncogenic action of 2-hydroxyglutarate | 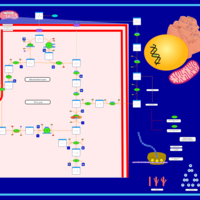 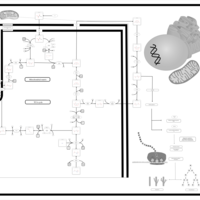 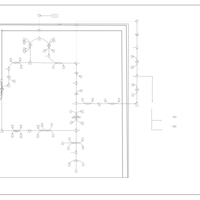 |
| The Oncogenic Action of Succinate | 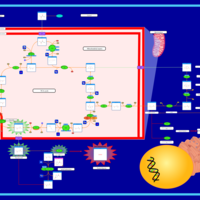  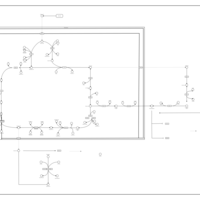 |
| The Oncogenic Action of Fumarate | 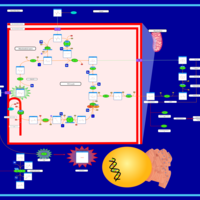 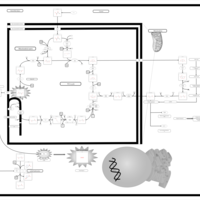  |
| Glutaminolysis and Cancer | 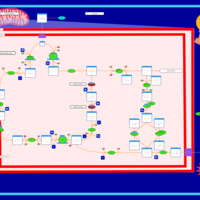 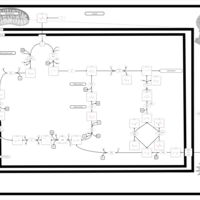 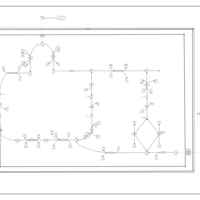 |
| The oncogenic action of L-2-hydroxyglutarate in Hydroxygluaricaciduria | 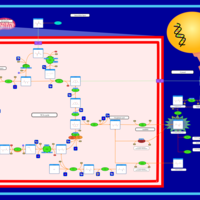 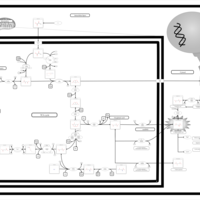 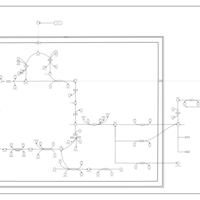 |
| The oncogenic action of D-2-hydroxyglutarate in Hydroxygluaricaciduria | 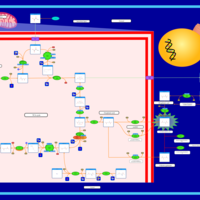 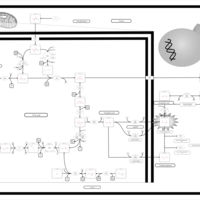 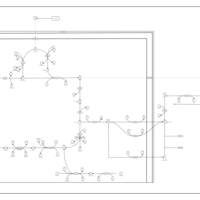 |
| Histamine H1 Receptor Activation |    |
| Chlorphenamine H1-Antihistamine Action | 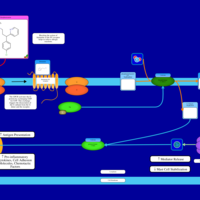   |
| Pheniramine H1-Antihistamine Action | 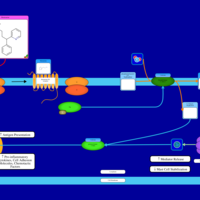 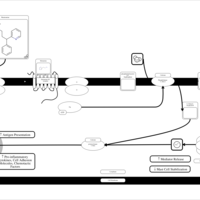  |
| Dexchlorpheniramine H1-Antihistamine Action | 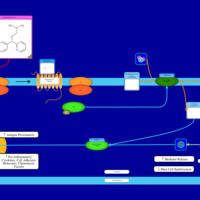 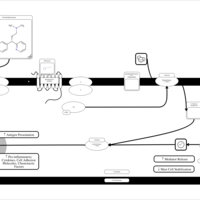 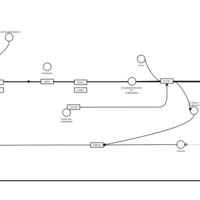 |
| Brompheniramine H1-Antihistamine Action | 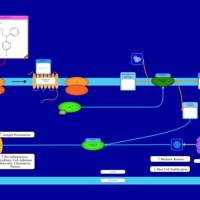 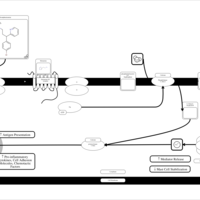 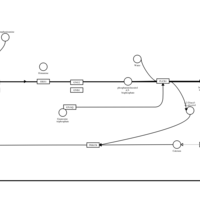 |
| Dexbrompheniramine H1-Antihistamine Action | 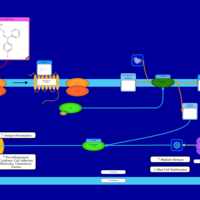 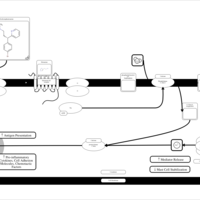  |
| Triprolidine H1-Antihistamine Action | 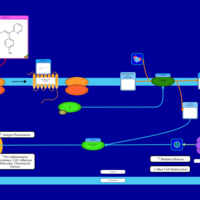 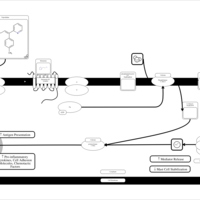  |
| Dimetindene H1-Antihistamine Action | 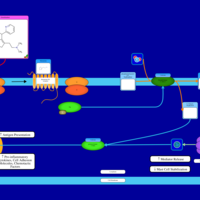 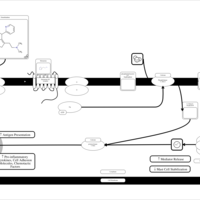  |
| Mepyramine H1-Antihistamine Action | 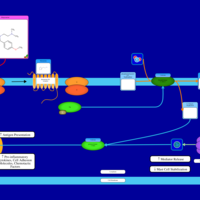 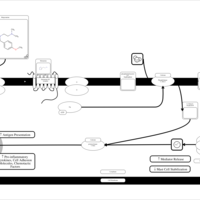  |
| Antazoline H1-Antihistamine Action | 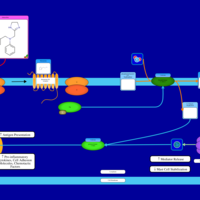 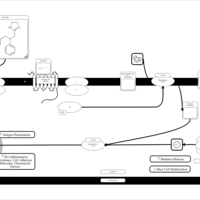  |
| Chloropyramine H1-Antihistamine Action |    |
| Talastine H1-Antihistamine Action | 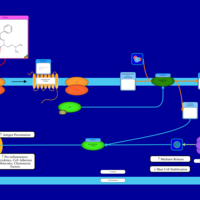 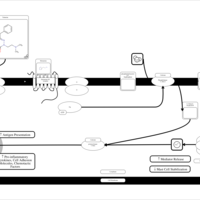 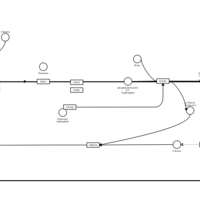 |
| Tripelennamine H1-Antihistamine Action | 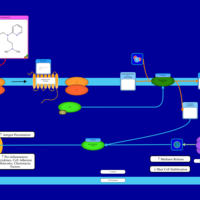 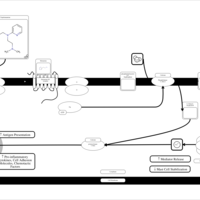 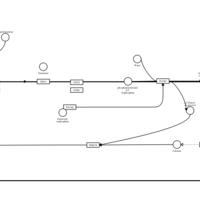 |
| Histapyrrodine H1-Antihistamine Action | 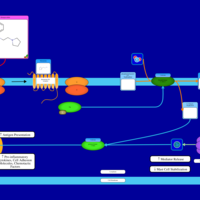 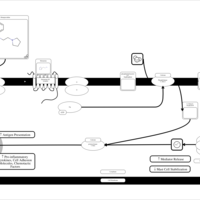 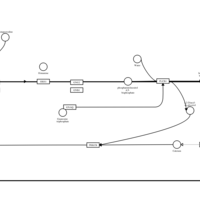 |
| Methapyrilene H1-Antihistamine Action |    |
| Thonzylamine H1-Antihistamine Action | 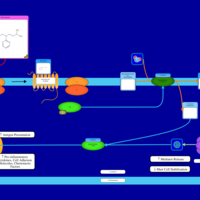 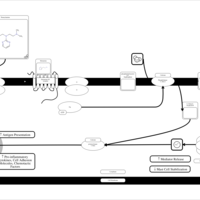  |
| Diphenhydramine H1-Antihistamine Action | 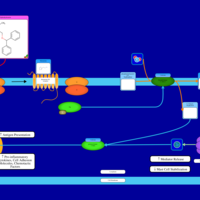 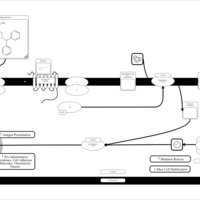 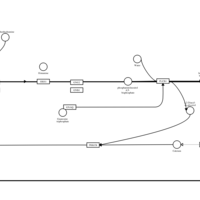 |
| Carbinoxamine H1-Antihistamine Action | 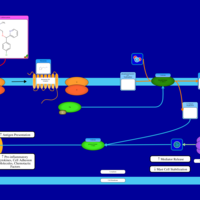 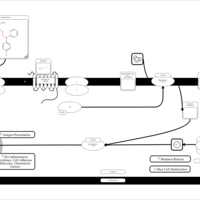 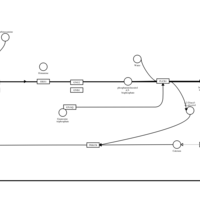 |
| Doxylamine H1-Antihistamine Action | 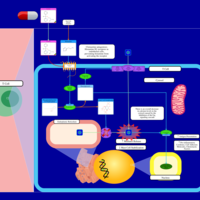   |
| Orphenadrine H1-Antihistamine Action | 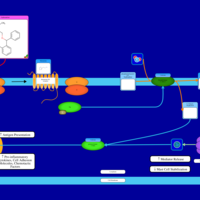 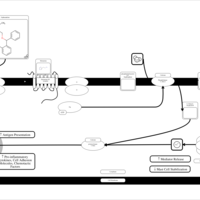  |
| Bromodiphenhydramine H1-Antihistamine Action | 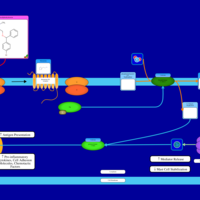 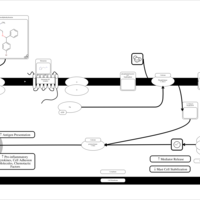  |
| Clemastine H1-Antihistamine Action | 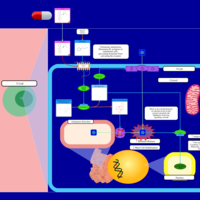   |
| Chlorphenoxamine H1-Antihistamine Action | 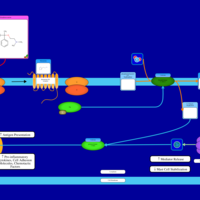 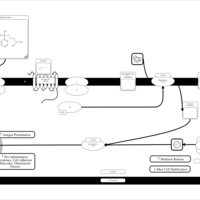 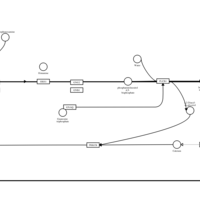 |
| Diphenylpyraline H1-Antihistamine Action | 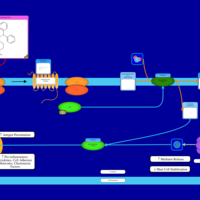 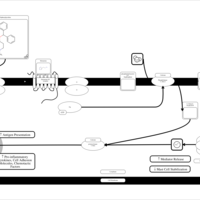 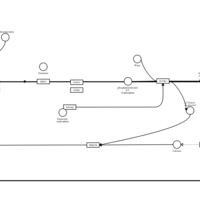 |
| Phenyltoloxamine H1-Antihistamine Action | 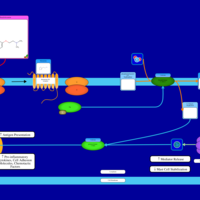 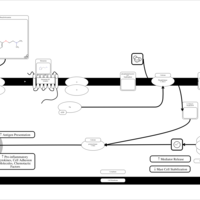 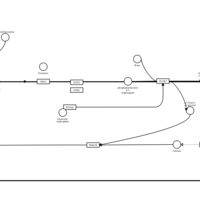 |
| Cyclizine H1-Antihistamine Action | 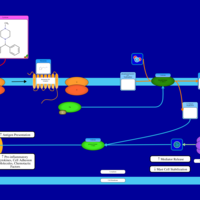   |
| Chlorcyclizine H1-Antihistamine Action | 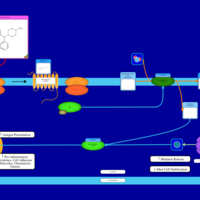 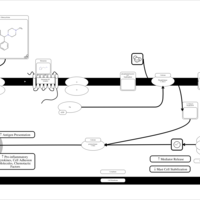 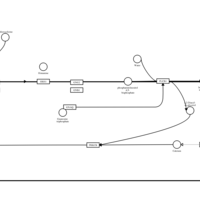 |
| Hydroxyzine H1-Antihistamine Action | 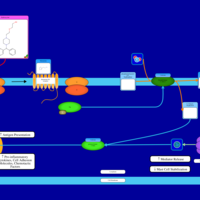  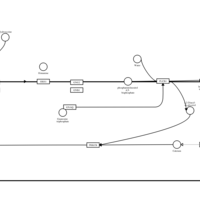 |
| Meclizine H1-Antihistamine Action | 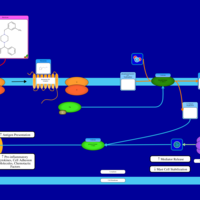   |
| Buclizine H1-Antihistamine Action | 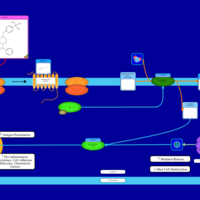 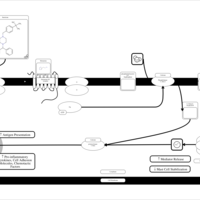  |
| Oxatomide H1-Antihistamine Action | 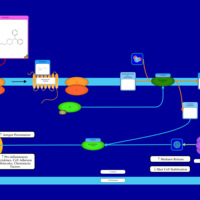 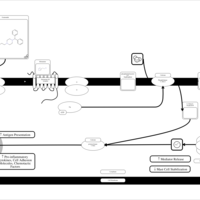 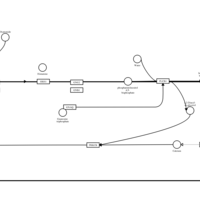 |
| Cetirizine H1-Antihistamine Action | 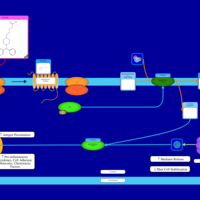 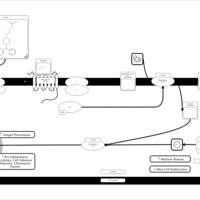 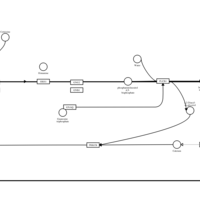 |
| Cinnarizine H1-Antihistamine Action | 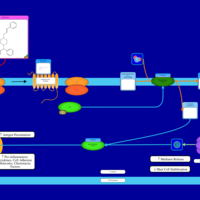 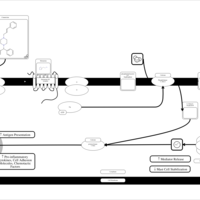  |
| Levocetirizine H1-Antihistamine Action | 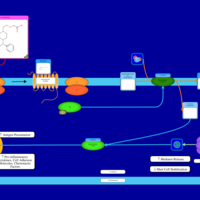 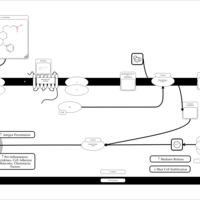  |
| Promethazine H1-Antihistamine Action | 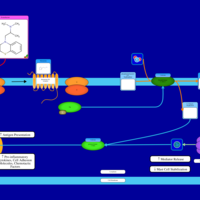 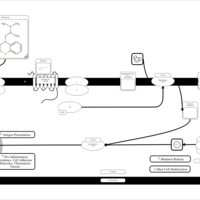 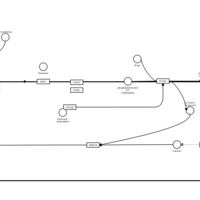 |
| Alimemazine H1-Antihistamine Action | 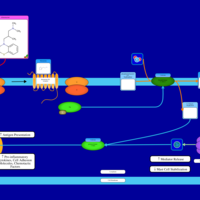 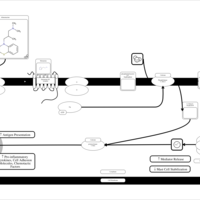 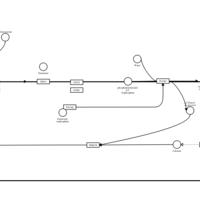 |
| Cyproheptadine H1-Antihistamine Action | 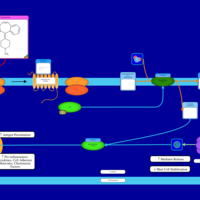 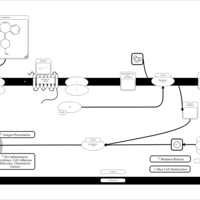  |
| Phenbenzamine H1-Antihistamine Action | 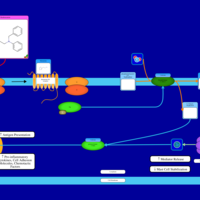 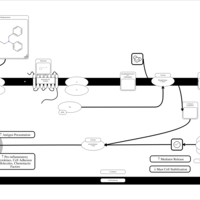  |
| Fenethazine H1-Antihistamine Action |    |
| Hydroxyethylpromethazine H1-Antihistamine Action | 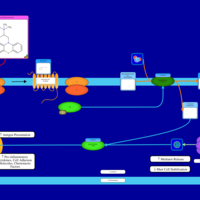 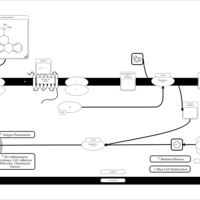 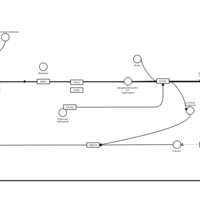 |
| Isothipendyl H1-Antihistamine Action | 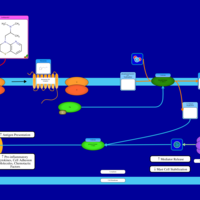 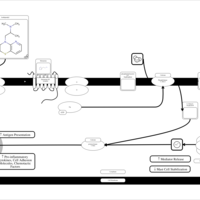  |
| Mequitazine H1-Antihistamine Action | 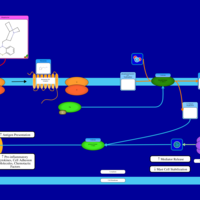 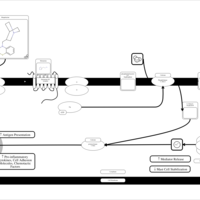  |
| Methdilazine H1-Antihistamine Action | 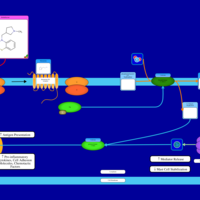 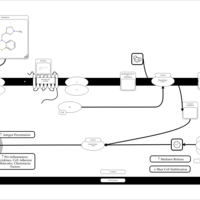 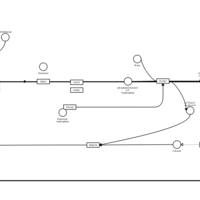 |
| Oxomemazine H1-Antihistamine Action | 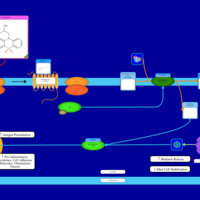  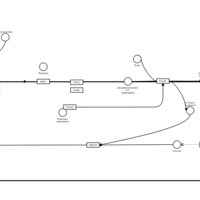 |
| Azatadine H1-Antihistamine Action | 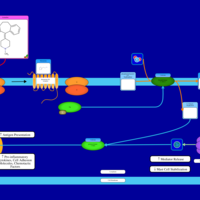 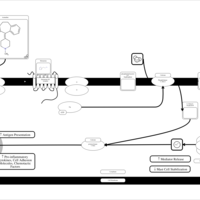  |
| Ketotifen H1-Antihistamine Action | 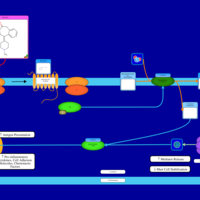 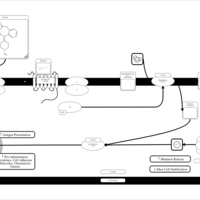 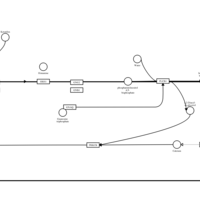 |
| Doxepin H1-Antihistamine Action | 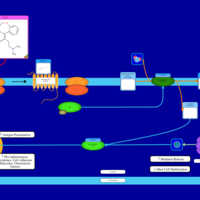 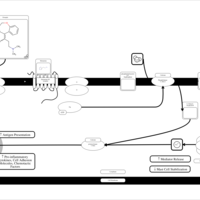 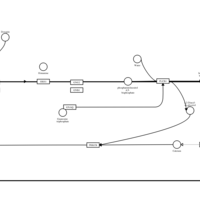 |
| Acrivastine H1-Antihistamine Action | 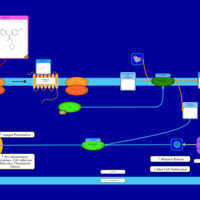  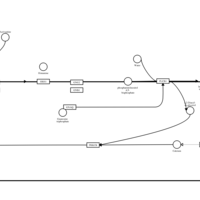 |
| Astemizole H1-Antihistamine Action | 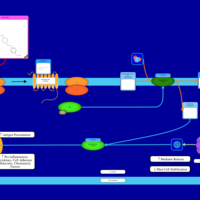 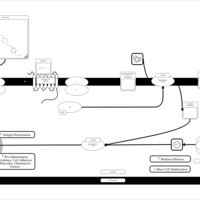  |
| Bepotastine H1-Antihistamine Action | 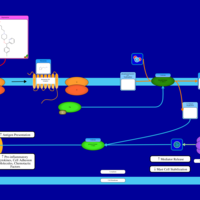 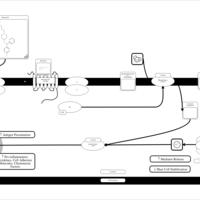  |
| Bilastine H1-Antihistamine Action | 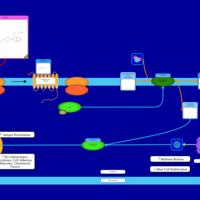  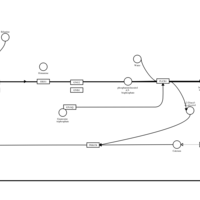 |
| Loratadine H1-Antihistamine Action | 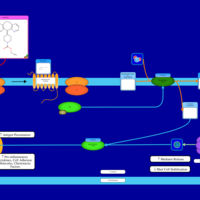 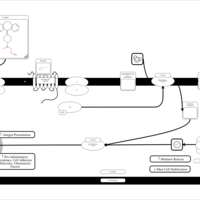 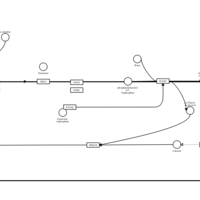 |
| Desloratadine H1-Antihistamine Action |   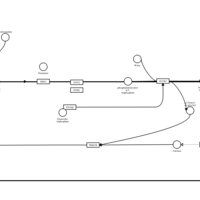 |
| Ebastine H1-Antihistamine Action | 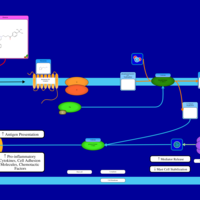 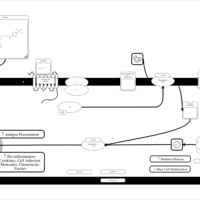 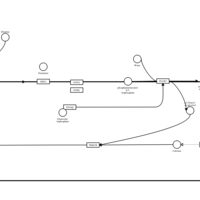 |
| Terfenadine H1-Antihistamine Action | 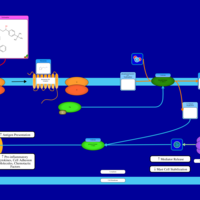 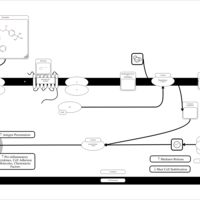  |
| Fexofenadine H1-Antihistamine Action | 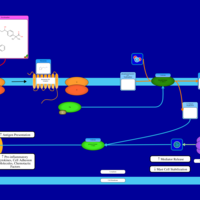 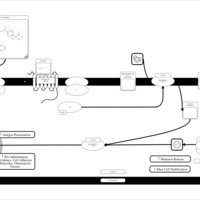 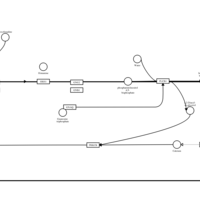 |
| Levocabastine H1-Antihistamine Action | 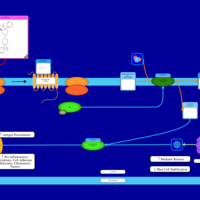 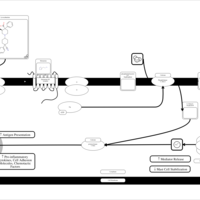 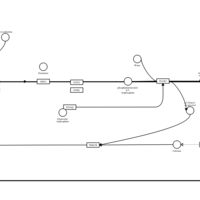 |
| Mizolastine H1-Antihistamine Action | 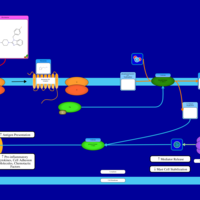 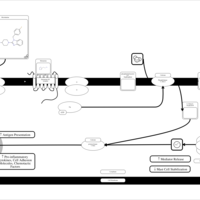 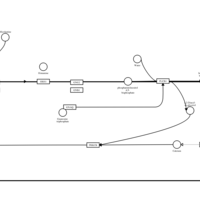 |
| Rupatadine H1-Antihistamine Action | 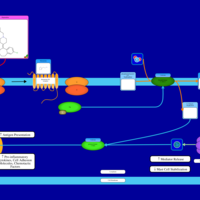 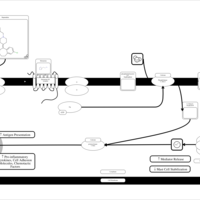  |
| Olopatadine H1-Antihistamine Action | 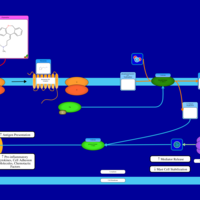 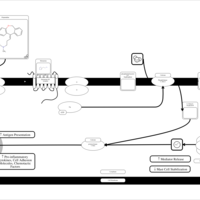 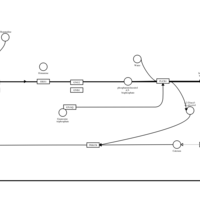 |
| Azelastine H1-Antihistamine Action | 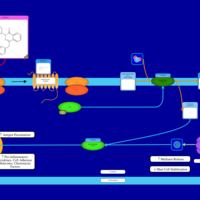 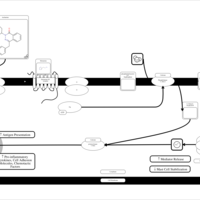 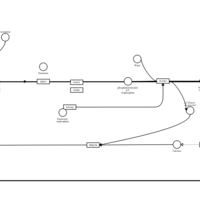 |
| Thiazinamium H1-Antihistamine Action | 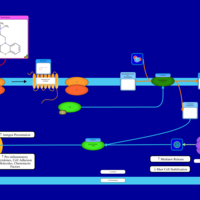 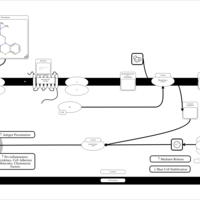 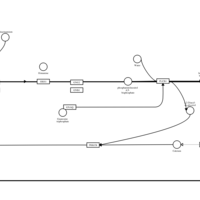 |
| Quifenadine H1-Antihistamine Action | 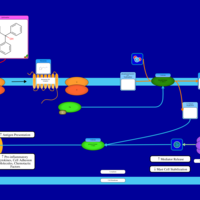 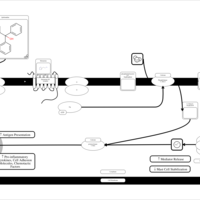  |
| Betahistine H1-Antihistamine Action | 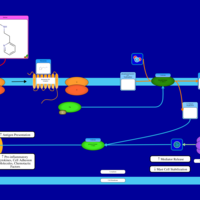 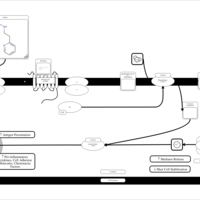 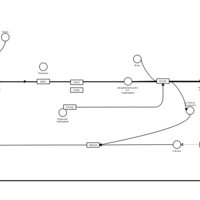 |
| Emedastine H1-Antihistamine Action | 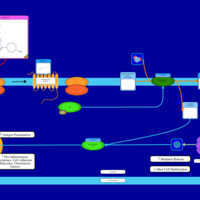 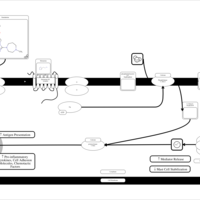 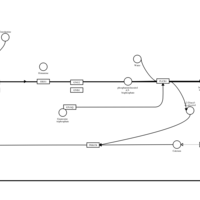 |
| Flunarizine H1-Antihistamine Action | 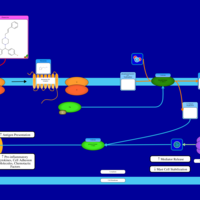  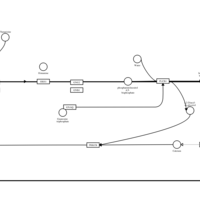 |
| Mebhydrolin H1-Antihistamine Action | 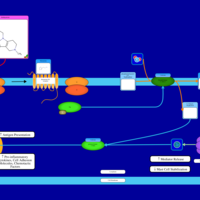 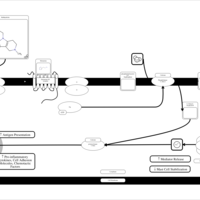 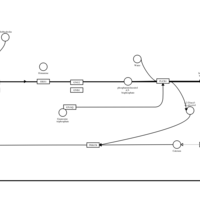 |
| Phenindamine H1-Antihistamine Action | 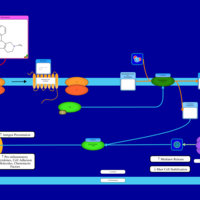 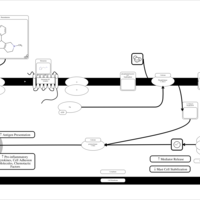 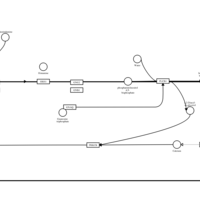 |
| Epinastine H1-Antihistamine Action | 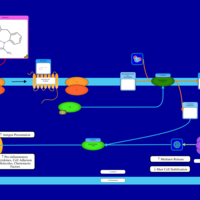  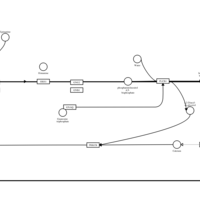 |
| Tolpropamine H1-Antihistamine Action | 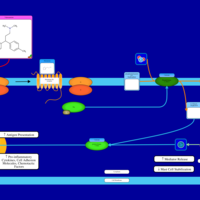 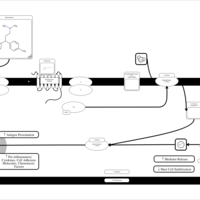  |
| Embramine H1-Antihistamine Action | 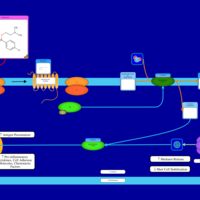 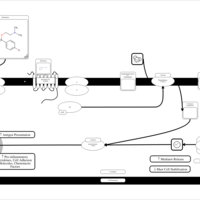  |
| Latrepirdine H1-Antihistamine Action | 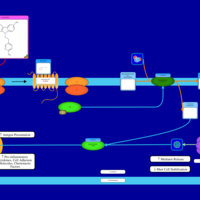  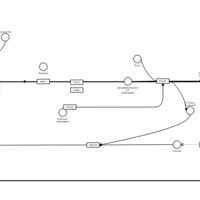 |
| Thenyldiamine H1-Antihistamine Action | 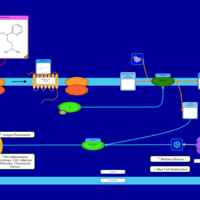 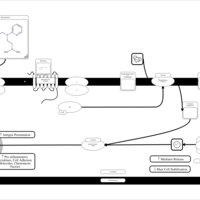 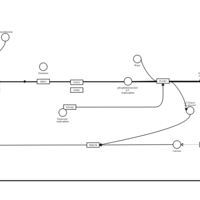 |
| Propiomazine H1-Antihistamine Action | 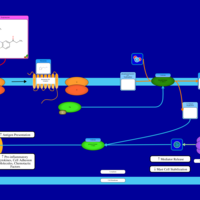 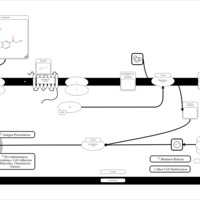 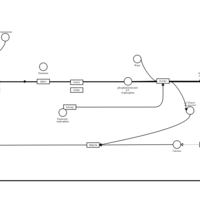 |
| Clocinizine H1-Antihistamine Action | 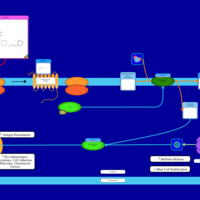 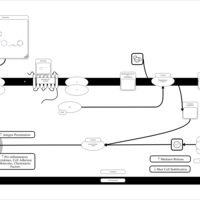  |
| Homochlorcyclizine H1-Antihistamine Action | 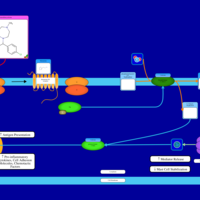 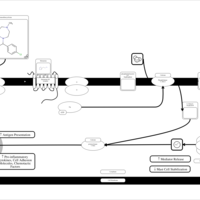 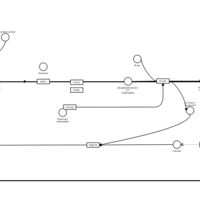 |
| Temelastine H1-Antihistamine Action | 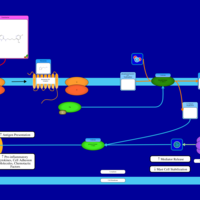 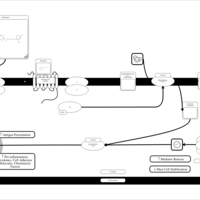 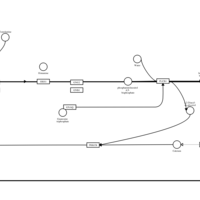 |
| Alcaftadine H1-Antihistamine Action | 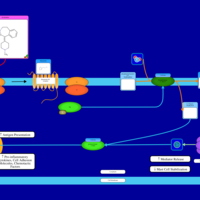   |
| Bamipine H1-Antihistamine Action | 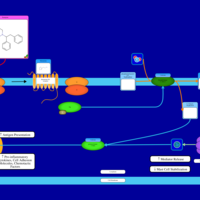 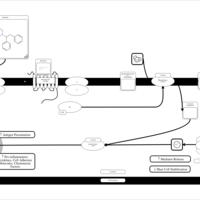  |
| Deptropine H1-Antihistamine Action | 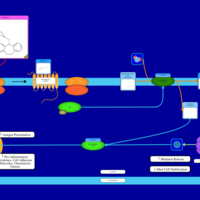 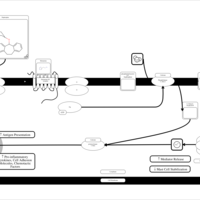 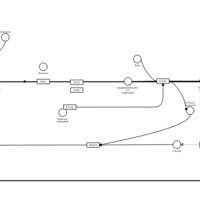 |
| Quetiapine H1-Antihistamine Action | 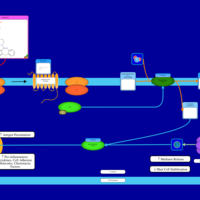 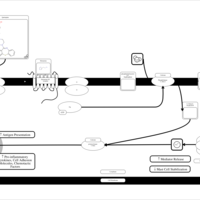 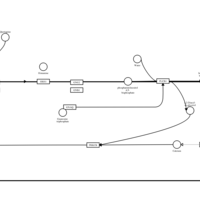 |
| Mirtazapine H1-Antihistamine Action | 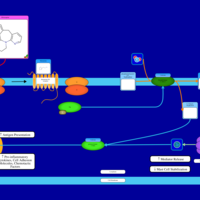   |
| Pimethixene H1-Antihistamine Action | 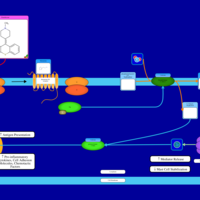 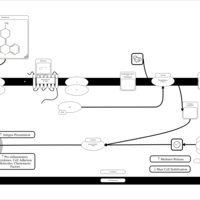  |
| Pyrrobutamine H1-Antihistamine Action | 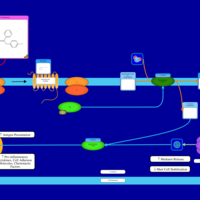 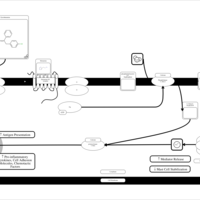 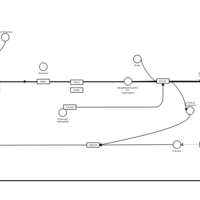 |
| Thenalidine H1-Antihistamine Action | 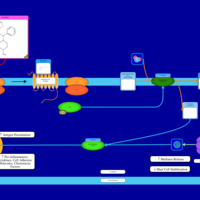 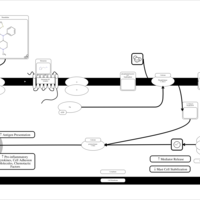 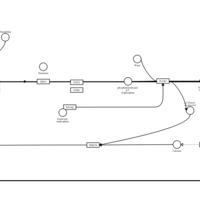 |
| Tritoqualine H1-Antihistamine Action |  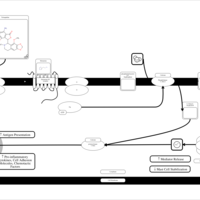 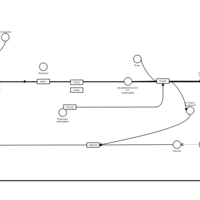 |
| Histamine H1 Receptor Activation |    |
| Lysophosphatidic Acid LPA1 Signalling | 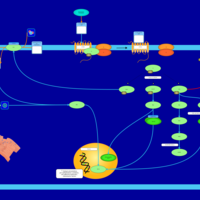 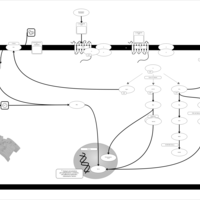 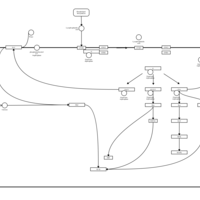 |
| Lysophosphatidic Acid LPA2 Signalling | 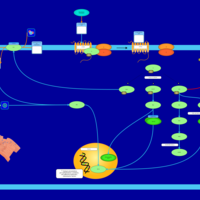 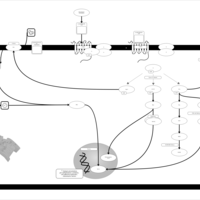 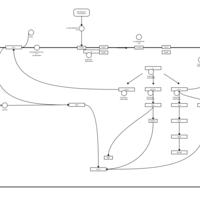 |
| Lysophosphatidic Acid LPA3 Signalling | 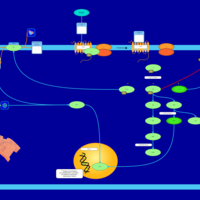 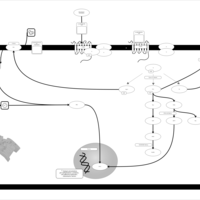 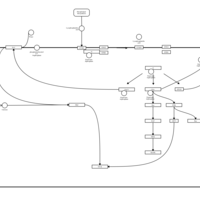 |
| Lysophosphatidic Acid LPA4 Signalling | 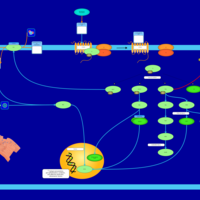 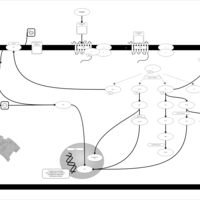  |
| Lysophosphatidic Acid LPA5 Signalling | 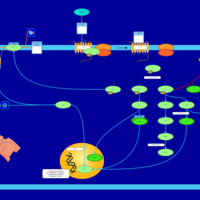 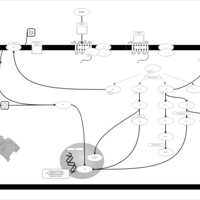 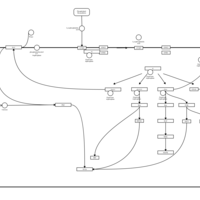 |
| Lysophosphatidic Acid LPA6 Signalling | 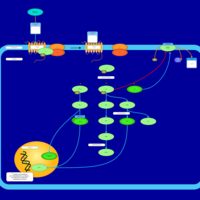 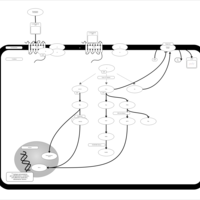 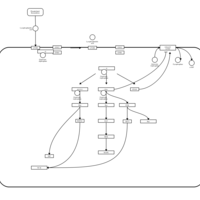 |