| Alanine, aspartate and glutamate metabolism | 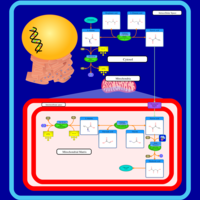 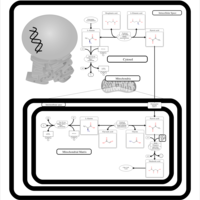  |
| Nitrogen metabolism |  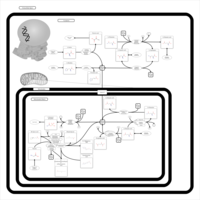  |
| Pyruvate metabolism |  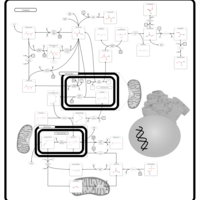  |
| Valine, leucine and isoleucine degradation |  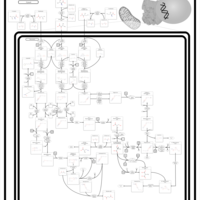 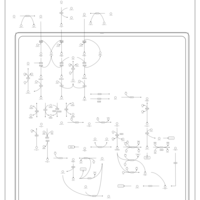 |
| Propanoate metabolism |  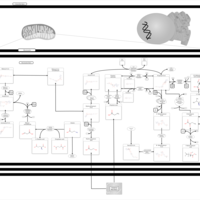 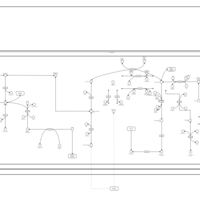 |
| Citric Acid Cycle | 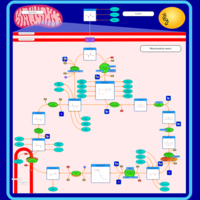 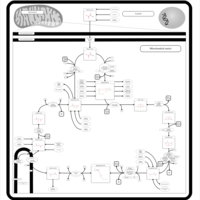 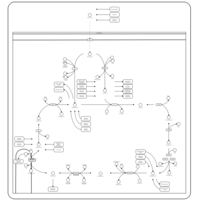 |
| Biotin Metabolism | 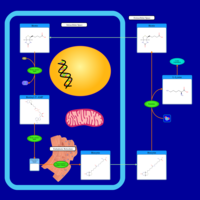 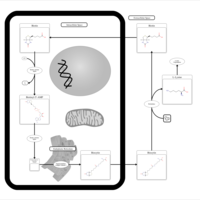 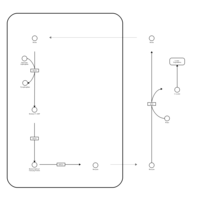 |
| Glutamate Metabolism | 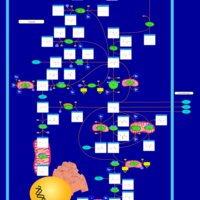 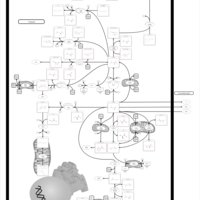 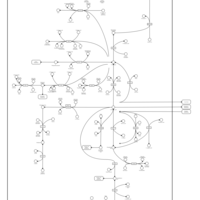 |
| Gluconeogenesis | 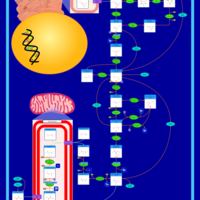 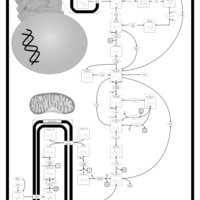 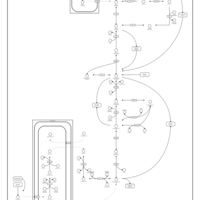 |
| 2-Hydroxyglutric Aciduria (D And L Form) | 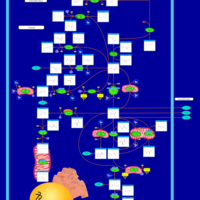 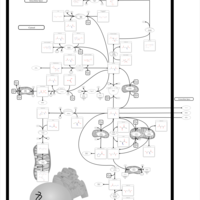 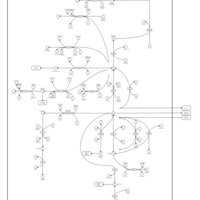 |
| 2-Methyl-3-Hydroxybutryl CoA Dehydrogenase Deficiency | 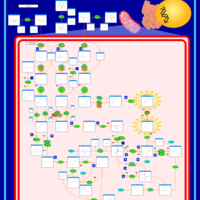 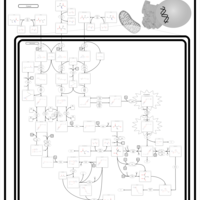 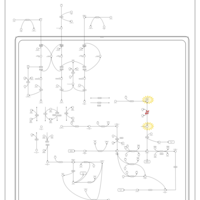 |
| 3-Hydroxy-3-Methylglutaryl-CoA Lyase Deficiency | 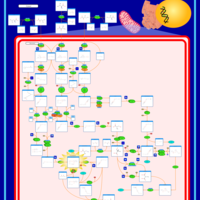 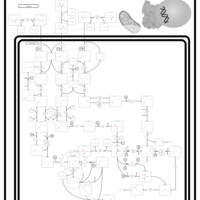 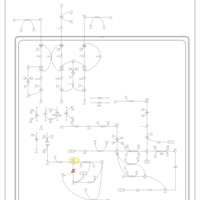 |
| 3-Methylglutaconic Aciduria Type I |  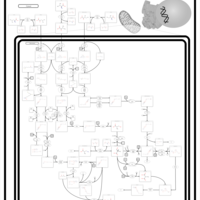 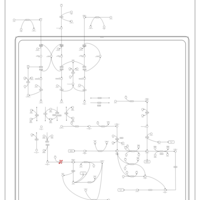 |
| 3-Methylglutaconic Aciduria Type III |  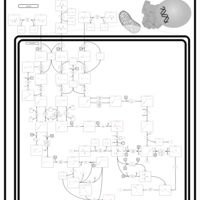 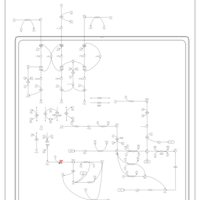 |
| 3-Methylglutaconic Aciduria Type IV | 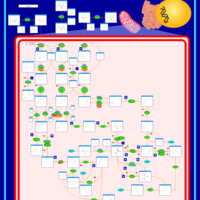 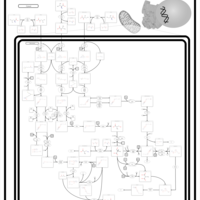  |
| Beta-Ketothiolase Deficiency | 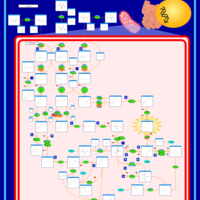 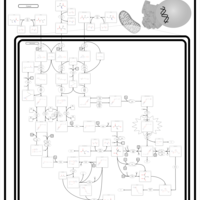 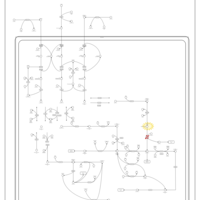 |
| Biotinidase Deficiency | 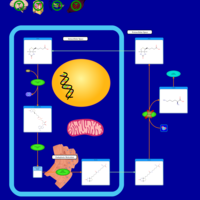 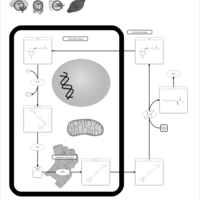 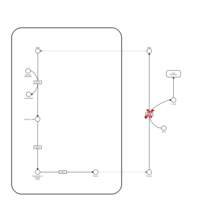 |
| Leigh Syndrome | 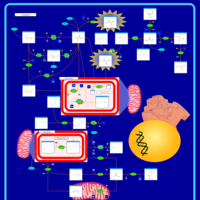 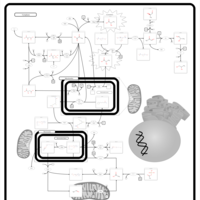 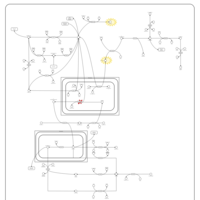 |
| Malonic Aciduria | 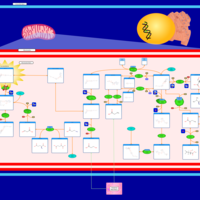  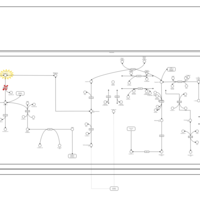 |
| Maple Syrup Urine Disease |    |
| Methylmalonic Aciduria |    |
| Methylmalonic Aciduria Due to Cobalamin-Related Disorders | 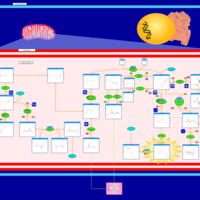 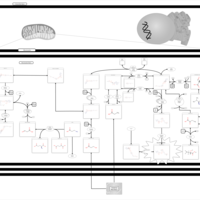 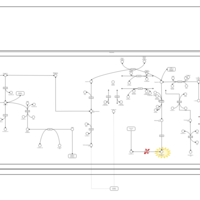 |
| Pyruvate Dehydrogenase Complex Deficiency | 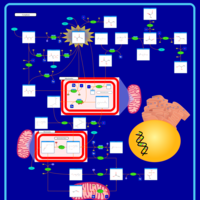 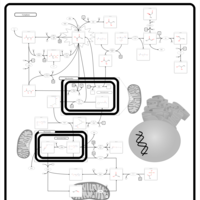 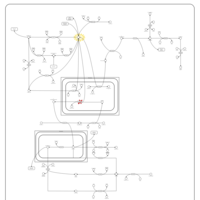 |
| Propionic Acidemia | 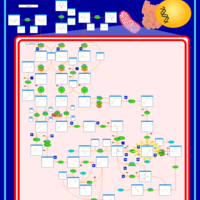 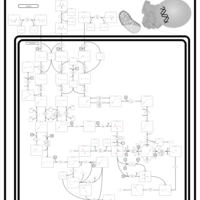  |
| 3-Methylcrotonyl Coa Carboxylase Deficiency Type I | 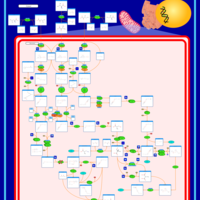 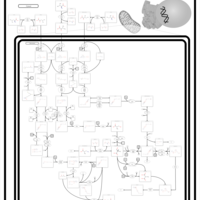 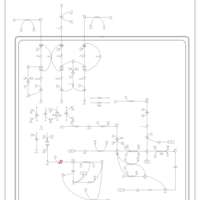 |
| Isovaleric Aciduria |    |
| 4-Hydroxybutyric Aciduria/Succinic Semialdehyde Dehydrogenase Deficiency | 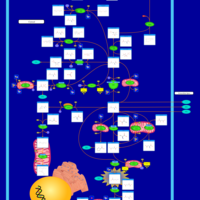 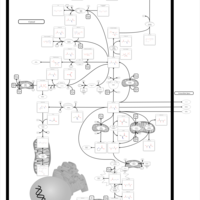 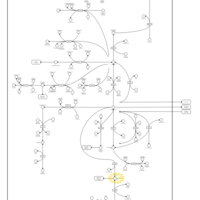 |
| Lactic Acidemia | 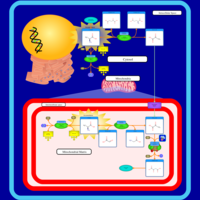 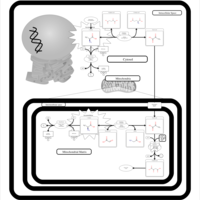 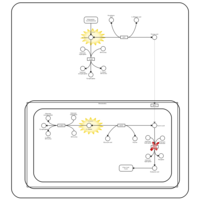 |
| Pyruvate Decarboxylase E1 Component Deficiency (PDHE1 Deficiency) | 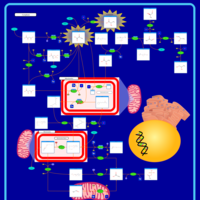 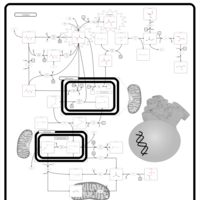 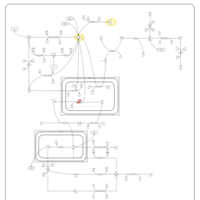 |
| Hyperinsulinism-Hyperammonemia Syndrome | 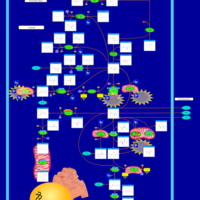 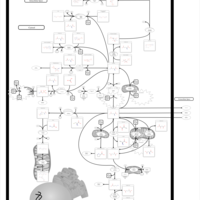 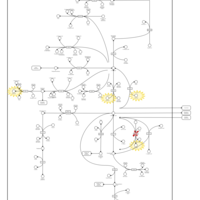 |
| Pyruvate Carboxylase Deficiency |  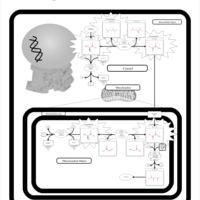 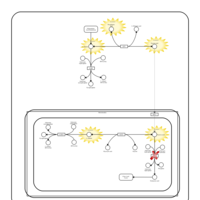 |
| Primary Hyperoxaluria Type I | 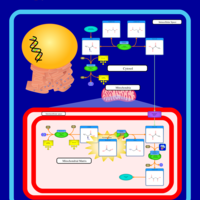 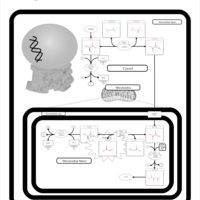 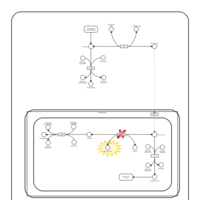 |
| Glycogen Storage Disease Type 1A (GSD1A) or Von Gierke Disease | 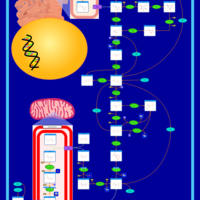 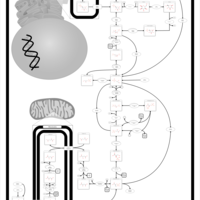 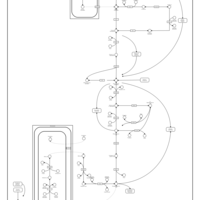 |
| Methylmalonate Semialdehyde Dehydrogenase Deficiency |    |
| Homocarnosinosis |   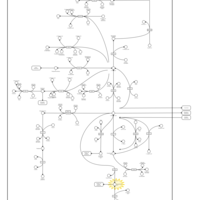 |
| Threonine and 2-Oxobutanoate Degradation | 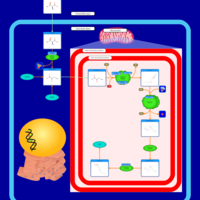 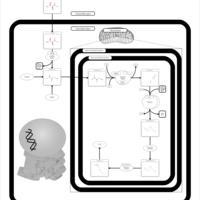 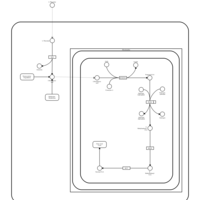 |
| Fatty Acid Biosynthesis | 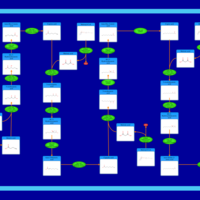 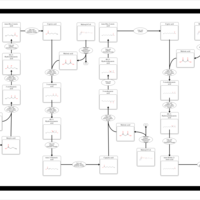 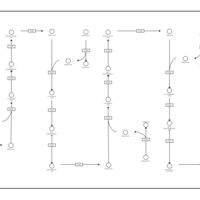 |
| Transfer of Acetyl Groups into Mitochondria | 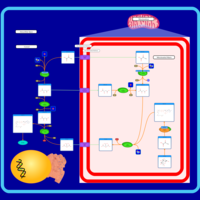 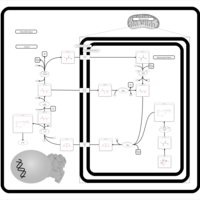 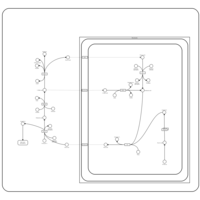 |
| Malonyl-coa decarboxylase deficiency | 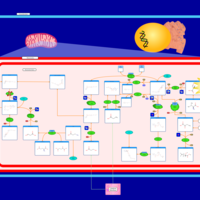 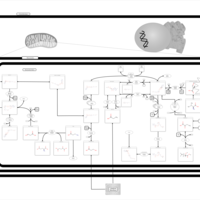 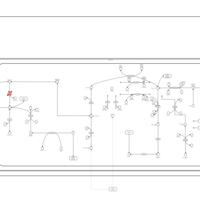 |
| 3-hydroxyisobutyric acid dehydrogenase deficiency |  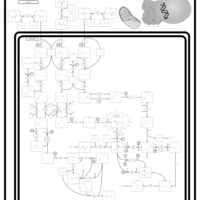 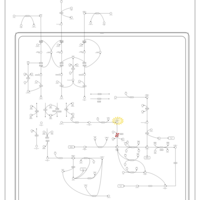 |
| 3-hydroxyisobutyric aciduria |  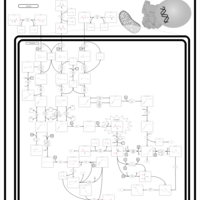 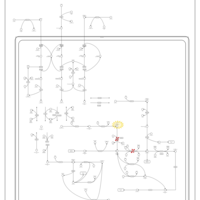 |
| Isobutyryl-coa dehydrogenase deficiency |    |
| Isovaleric acidemia |    |
| Congenital lactic acidosis | 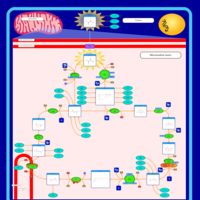 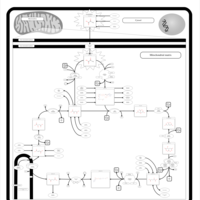 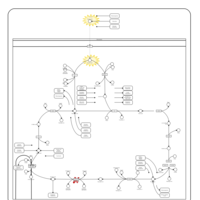 |
| Fumarase deficiency |  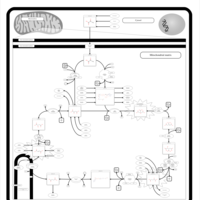 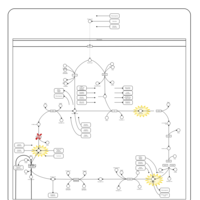 |
| Mitochondrial complex II deficiency | 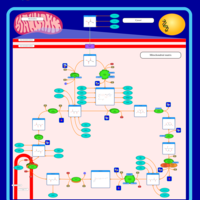 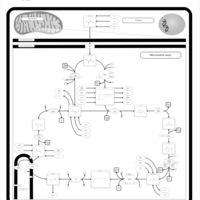 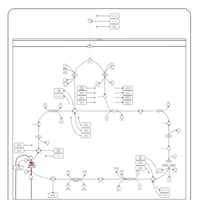 |
| 2-ketoglutarate dehydrogenase complex deficiency | 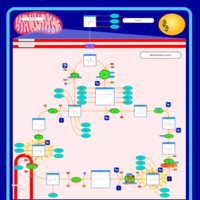 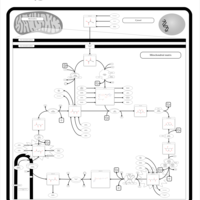 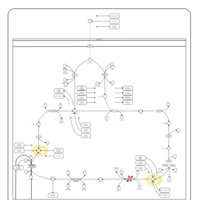 |
| Pyruvate dehydrogenase deficiency (E3) |  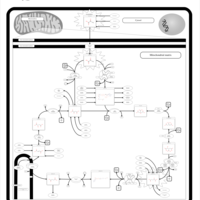 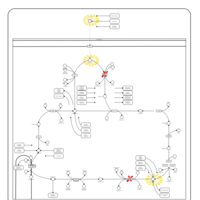 |
| Pyruvate dehydrogenase deficiency (E2) |  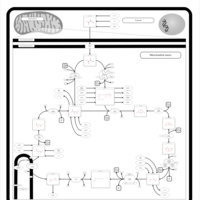 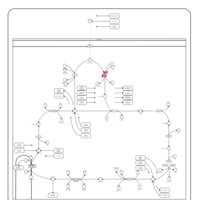 |
| Primary hyperoxaluria II, PH2 | 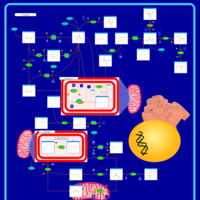 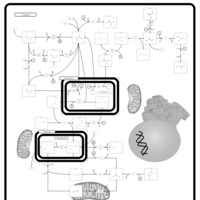  |
| Pyruvate kinase deficiency | 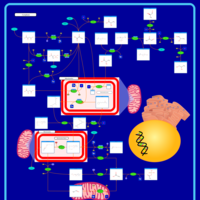 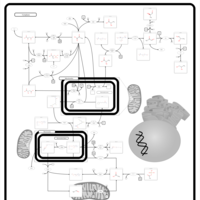 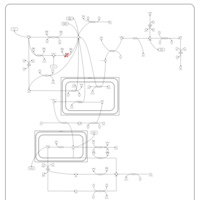 |
| Phosphoenolpyruvate carboxykinase deficiency 1 (PEPCK1) | 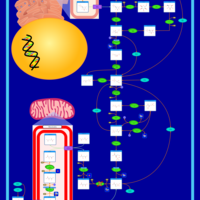 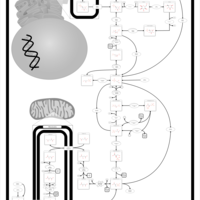 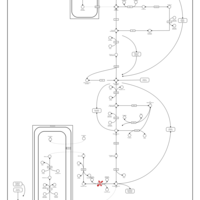 |
| Fructose-1,6-diphosphatase deficiency | 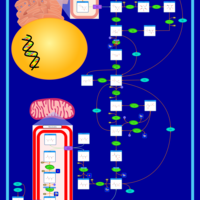 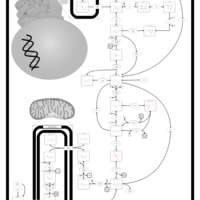 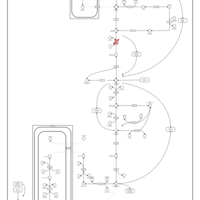 |
| Triosephosphate isomerase | 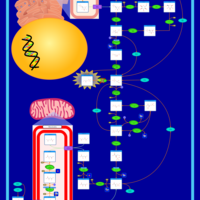 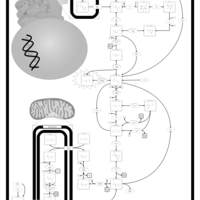 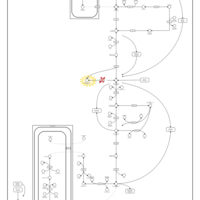 |
| Multiple carboxylase deficiency, neonatal or early onset form | 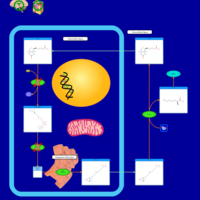 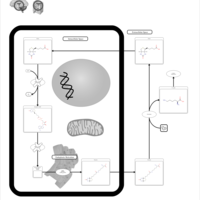  |
| Succinic semialdehyde dehydrogenase deficiency | 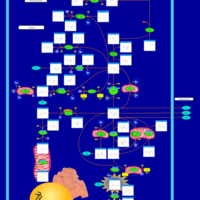 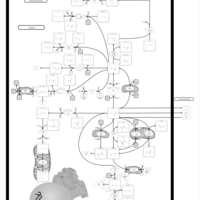 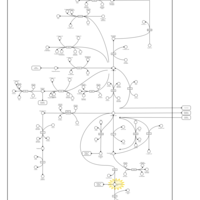 |
| Glycogenosis, Type IB |    |
| Glycogenosis, Type IC |    |
| Glycogenosis, Type IA. Von gierke disease |    |
| Warburg Effect | 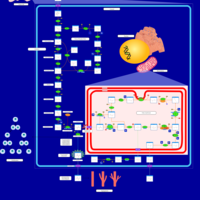 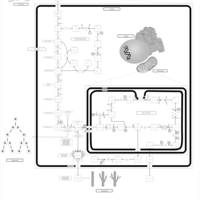 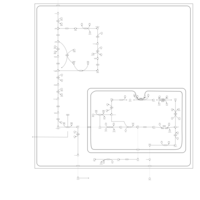 |
| Acylcarnitine 3-Hydroxydecanoylcarnitine |  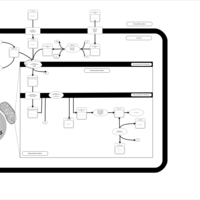  |
| Acylcarnitine (7Z,9Z,12Z,15Z,18Z,21Z)-tetracosa-7,9,12,15,18,21-hexaenoylcarnitine | 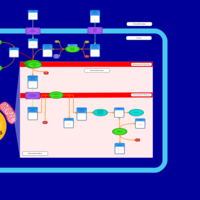 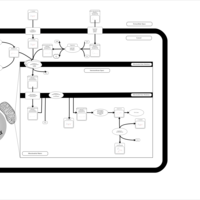 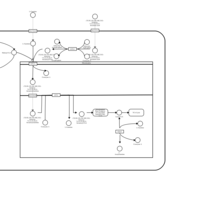 |
| Acylcarnitine Pentacosanoylcarnitine | 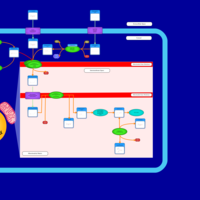 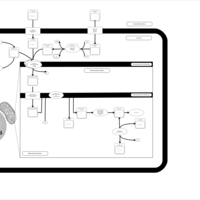 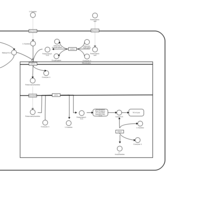 |
| Acylcarnitine Hexacosanoylcarnitine | 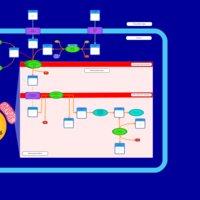 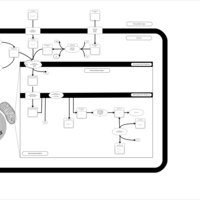 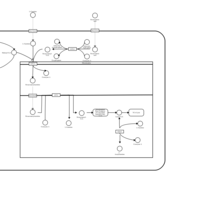 |
| Acylcarnitine (17Z)-Hexacos-17-enoylcarnitine | 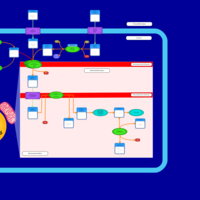 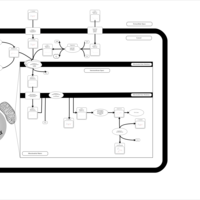  |
| Acylcarnitine (13Z,16Z)-Hexacosa-13,16-dienoylcarnitine |    |
| Acylcarnitine Heptacosanoylcarnitine |  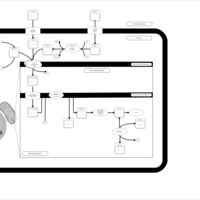 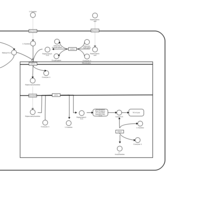 |
| Acylcarnitine Octacosanoylcarnitine | 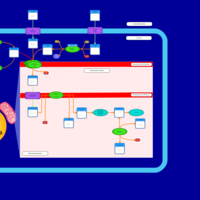 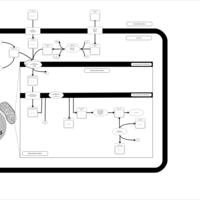 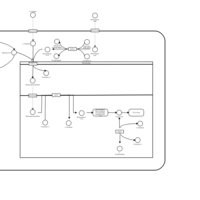 |
| Acylcarnitine Nonacosanoylcarnitine | 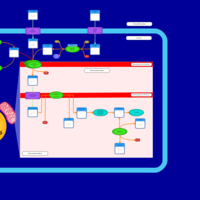 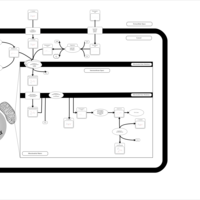 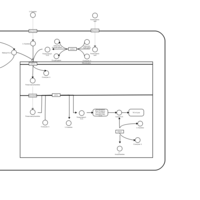 |
| Acylcarnitine (2S,3R)-3-Hydroxy-2-methylbutanoylcarnitine | 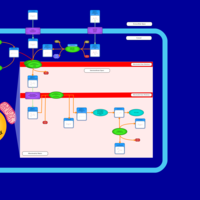 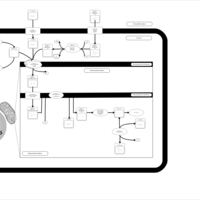 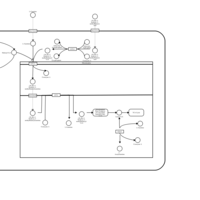 |
| The oncogenic action of 2-hydroxyglutarate | 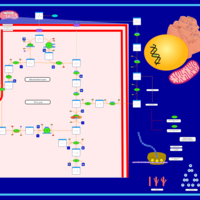 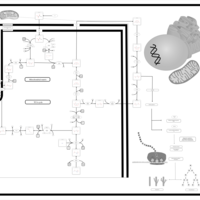 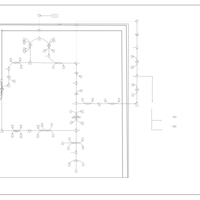 |
| The Oncogenic Action of Succinate | 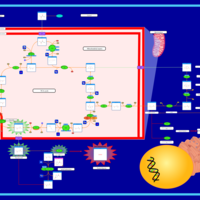  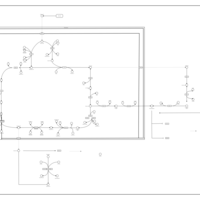 |
| The Oncogenic Action of Fumarate | 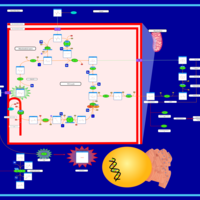 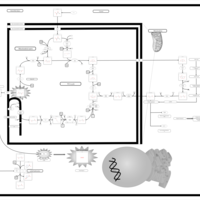  |
| Glutaminolysis and Cancer | 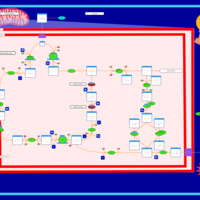 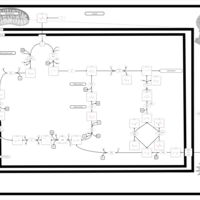 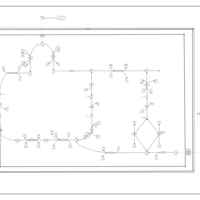 |
| The oncogenic action of L-2-hydroxyglutarate in Hydroxygluaricaciduria | 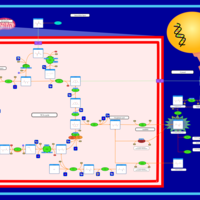 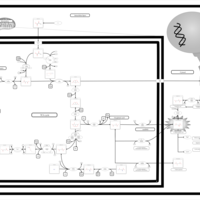 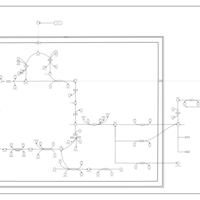 |
| The oncogenic action of D-2-hydroxyglutarate in Hydroxygluaricaciduria | 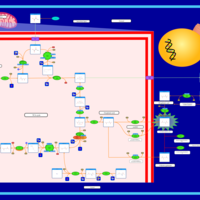 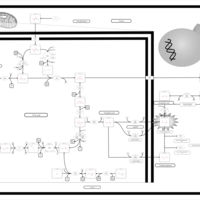 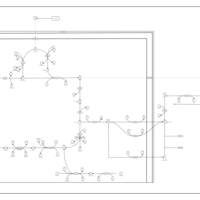 |